Difference between revisions of "Part:BBa C0161:Experience"
(→Induction by the Supernatant of a LuxI Producing Culture) |
(→Results) |
||
| (20 intermediate revisions by 3 users not shown) | |||
| Line 4: | Line 4: | ||
===Applications of BBa_C0161=== | ===Applications of BBa_C0161=== | ||
| + | |||
| + | ===Characterization of BBa_C0161-Arizona_State 2016=== | ||
| + | Authors: Ernesto Luna, Brady Dennison, Cassandra Barrett, Jimmy Xu, Jiaqi Wu, Dr. Karmella Haynes | ||
| + | |||
| + | Our team helped increase characterization of the part BBa_C0161 (LuxI). This part was tested against its ability to induce the part BBa_F2620 by the Canton Lab(MIT). This part outputs PoPS as a Receiver Device combined with LuxR. An induction test on BBa_F2620 had been done by Dr. Barry Canton (2008), but they tested GFP production over various AHL concentrations, while our test was an 8-hour GFP read over time for 2 AHL concentrations (10 and 50%). In addition, the Canton test utilized synthetic 3-O-C6-HSLs while our test utilized AHLs produced via an E.coli chassis. A visual induction test was also done, plating the Sender alongside a GFP positive control, negative receiver control, and F2620. | ||
| + | |||
| + | As shown below, Lux is able to induce F2620, as some colonies in the top right section began producing GFP. This is the expected result, since the Receiver of this system naturally accepts 3-O-C6-HSLs. | ||
| + | <div style="text-align: center;">[[File:T--Arizona_State--luxplate.png]]</div> | ||
| + | <div style="text-align: center;">Plate with GFP+(top left), Sender(center), -Receiver(bottom) and F2620(top right)</div> | ||
| + | |||
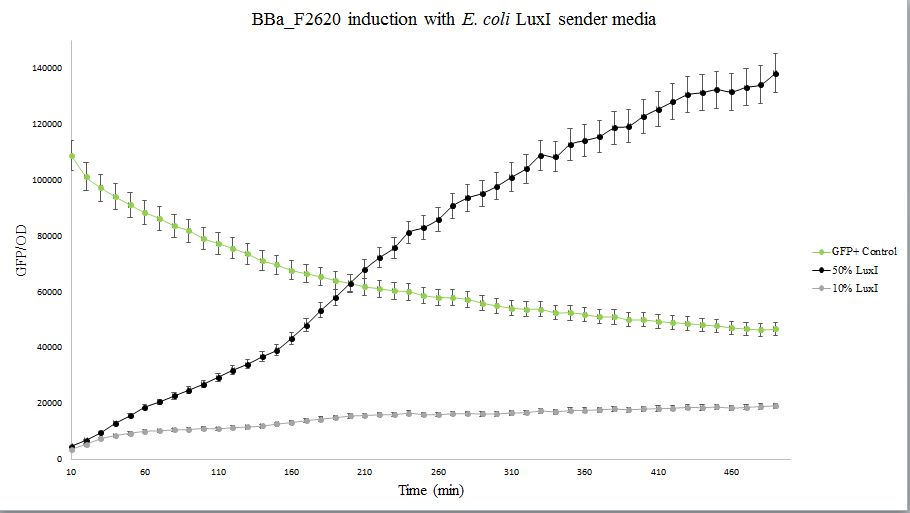
| + | The figure below compares LuxI at 10% and 50% concentrations. LuxI is shown to induce F2620. This affirms that F2620 is capable of being induced by LuxI synthesized within BL21(DE3) E. coli. | ||
| + | <div style="text-align: center;">[[File:T--Arizona_State--luxind.png]]</div> | ||
| + | <div style="text-align: center;">GFP absorbance from LuxI over time</div> | ||
| + | |||
| + | ====AHL Disposal Test==== | ||
| + | |||
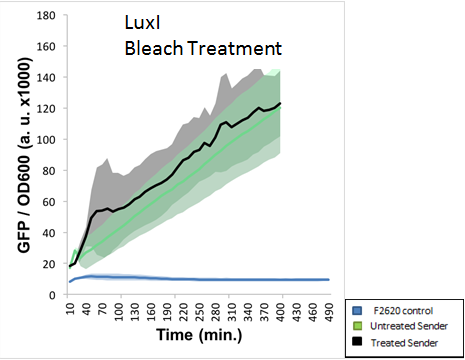
| + | The final experiment conducted using this part aimed to determine proper safe disposal procedures for the 3-O-C6-HSL. This AHL molecule is capable of crosstalk with potentially pathogenic strains of bacteria, and proper disposal of these AHLs should be an important biosafety measure taken. S.A. Borchardt had already tested the susceptibility of AHLs to bleach and found that 3-oxo AHLs were easily broken down by bleach while other AHLs were not. Our experiment aimed to test the application of standard EH&S sanitation protocols on AHLs (10% bleach solution and autoclaving). The figure below indicates that AHLs produced by LuxI were not properly deactivated by a 10% bleach solution. This was NOT the expected result, as LuxI produces a 3-oxo AHL, which should have been destroyed by bleach. | ||
| + | <div style="text-align: center;">[[File:T--Arizona_State--luxbleachgraph.png]]</div> | ||
| + | <div style="text-align: center;">GFP absorbance from LuxI over time</div> | ||
| + | |||
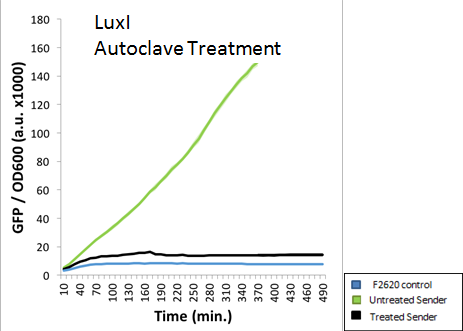
| + | A standard 15 minute Liquid autoclave cycle was also used to treat an extracted AHL solution. The figure below indicates that LuxI was nearly completely destroyed via autoclaving. This was the expected result, as the high pressure and temperatures should deactivate any AHL molecules present. | ||
| + | <div style="text-align: center;">[[File:T--Arizona State--luxautoclavegraph1.png]]</div> | ||
| + | <div style="text-align: center;">GFP absorbance from LuxI over time</div> | ||
| + | |||
| + | ====Conclusion==== | ||
| + | |||
| + | The results demonstrate that Lux was able to effectively induce F2620 after being extracted. The Lux results were not consistent, which showed decreased induction when treated with bleach, but no evidence of complete AHL inactivation. According to the autoclave results, a standard 15 min liquid procedure is able to degrade nearly all AHLs. The extreme pressure and temperature generated by the autoclave was more than enough to remove any threat posed by these AHL samples. In summary, our data suggests that autoclaving will ensure at least some inactivation of AHLs, while bleach may have no effect at all on the degradation of AHLs. | ||
===User Reviews=== | ===User Reviews=== | ||
| Line 30: | Line 57: | ||
== Background information == | == Background information == | ||
| − | [[Part:BBa_C0161|LuxI]] synthesizes [[3OC6HSL|3OC6-HSL]] which can bind to the regulator [[Part:BBa_C0062|LuxR]]. The [[Part:BBa_C0062|LuxR]]/[[3OC6HSL|3OC6-HSL]] complex can | + | [[Part:BBa_C0161|LuxI]] synthesizes [[3OC6HSL|3OC6-HSL]] which can bind to the regulator [[Part:BBa_C0062|LuxR]]. The [[Part:BBa_C0062|LuxR]]/[[3OC6HSL|3OC6-HSL]] complex can subsequently induce the promoter [[Part:BBa_R0062|pLux]]. |
In this experiment we used ''E. coli'' TOP10 strain transformed with a plasmid containing a pBBR1 origin of replication and a tetracycline resistance gene, the constitutive promoter [[Part:BBa_J23100|J23100]] from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson collection], an optimized RBS for this genetic context and the gene for the [[3OC6HSL|3OC6-HSL]] synthetase [[Part:BBa_C0161|LuxI]]. The [[Ribosome_Binding_Sites|RBS]] was optimized using the [http://salis.psu.edu/software/RBSLibraryCalculatorSearchMode Salis Lab RBS Calculator]. This strain was used as a [[3OC6HSL|3OC6-HSL]] producer strain. The detailed construct design and full sequence (piG0050max) is [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. | In this experiment we used ''E. coli'' TOP10 strain transformed with a plasmid containing a pBBR1 origin of replication and a tetracycline resistance gene, the constitutive promoter [[Part:BBa_J23100|J23100]] from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson collection], an optimized RBS for this genetic context and the gene for the [[3OC6HSL|3OC6-HSL]] synthetase [[Part:BBa_C0161|LuxI]]. The [[Ribosome_Binding_Sites|RBS]] was optimized using the [http://salis.psu.edu/software/RBSLibraryCalculatorSearchMode Salis Lab RBS Calculator]. This strain was used as a [[3OC6HSL|3OC6-HSL]] producer strain. The detailed construct design and full sequence (piG0050max) is [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. | ||
| − | As sensor strain we used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter [[Part:BBa_R0062|pLux]] within a [http://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator system] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [[Part:BBa_B0040|BBa_B0040]] and [[Part:BBa_B0015|BBa_B0015]] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and one of three promoters chosen from the Anderson promoter collection followed by [[Part: | + | As sensor strain we used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter [[Part:BBa_R0062|pLux]] within a [http://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator system] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [[Part:BBa_B0040|BBa_B0040]] and [[Part:BBa_B0015|BBa_B0015]] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and one of three promoters chosen from the Anderson promoter collection followed by [[Part:BBa_C0062|LuxR]]. A third plasmid containing the pBBR1 origin and a tetracycline resistance was introduced, since the [[Part:BBa_C0161|LuxI]] expressing [[3OC6HSL|3OC6-HSL]]-producer strain contained a tetracycline resistance an therefore the supernatant also contained tetracycline. The detailed construct designs and full sequences (piG0041, piG0065) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. |
== Experimental Setup == | == Experimental Setup == | ||
| Line 59: | Line 86: | ||
This review comes from the old result system and indicates that this part did not work in some test. | This review comes from the old result system and indicates that this part did not work in some test. | ||
|} | |} | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | =ShanghaiTech_China 2022's Characterization= | ||
| + | |||
| + | ===Design=== | ||
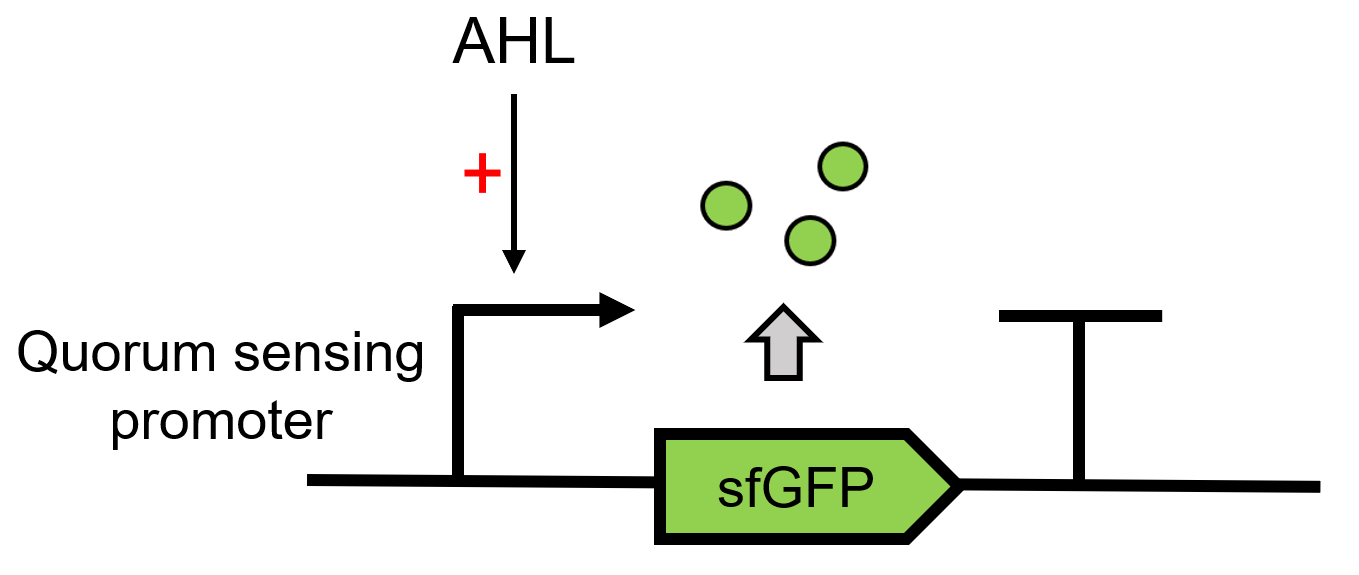
| + | LuxI can send AHL, 3OC6 HSL here and the signal can be received by the LuxR[https://parts.igem.org/Part:BBa_C0062 (BBa_C0062)]. LuxR can then regulate the downstream Lux pR [https://parts.igem.org/Part:BBa_R0062 (BBa_R0062)] to make superfolder GFP [https://parts.igem.org/Part:BBa_K4115000 (BBa_K4115000)] express.(Figure 1)<br> | ||
| + | |||
| + | [[File:Results-QS report.png|550px|thumb|left|'''Figure 1:''' Lux QS system path.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | ===Results=== | ||
| + | We use LuxI supernatant to induce LuxR to respond and express sfGFP.The test group is compared with 10<sup>-7</sup>M 3OC6 HSL induced group and no induced group(blank group). The result is shown by dividing the fluorescence intensity by the OD600, both measured in the transformed <i>E. coil</i>. (Figure 2) | ||
| + | |||
| + | [[File:LuxI supernatant induces LuxR.jpg|500px|thumb|left|'''Figure 2:''' LuxI supernatant induces LuxR to respond and express sfGFP.The data is compared with 10<sup>-7</sup>M 3OC6 HSL added group and the blank group is the <i>E. coli</i> culture.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
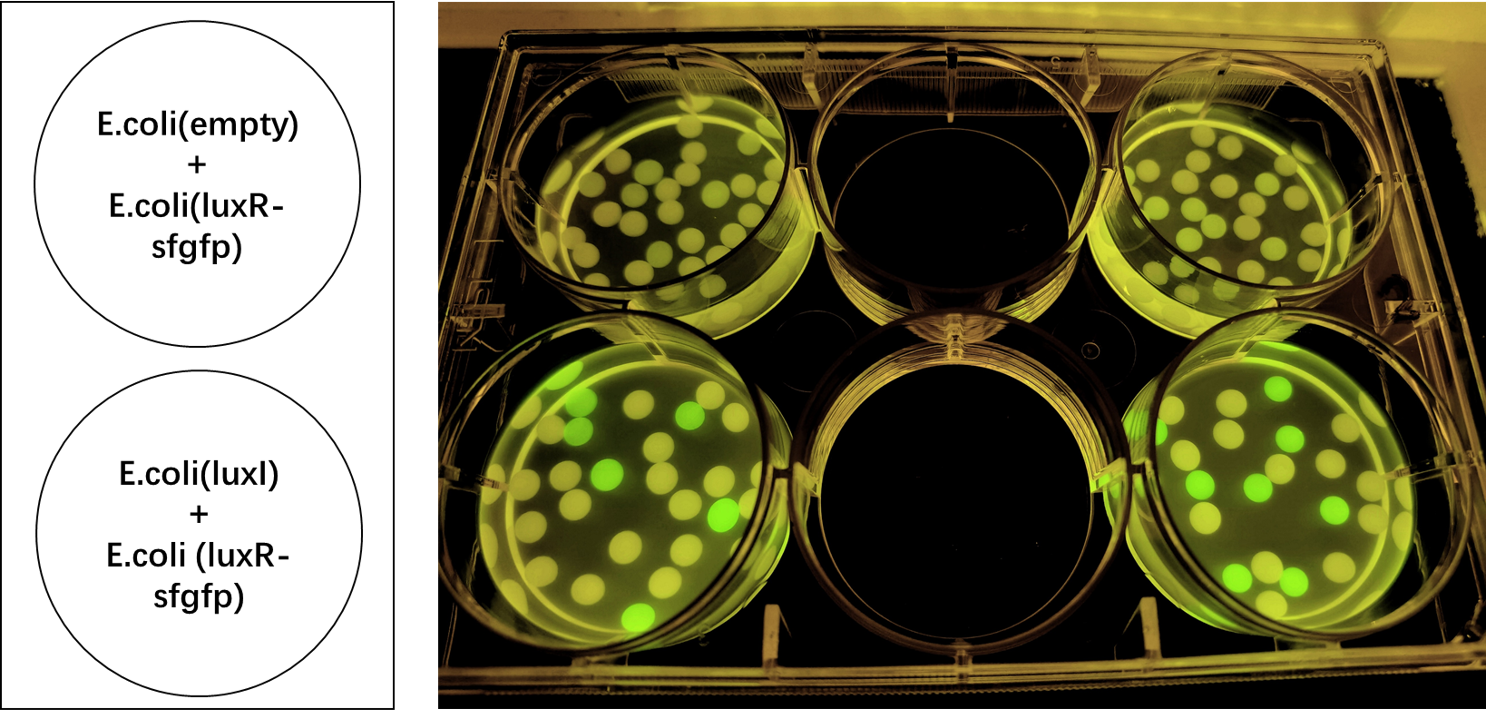
| + | We also induced the expression of LuxR in sodium alginate gel beads and detected the expression of sfGFP under UV light. (Figure 3)<br> | ||
| + | |||
| + | [[File:LuxR regulations between E.coli.jpg|700px|thumb|left|'''Figure 3:''' sfGFP is expressed under the regulation of LuxR between <i>E. coli</i> in a single gel bead. Two parallel groups are given and the above two are control groups.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | More detailed information can be seen on the composite page [https://parts.igem.org/Part:BBa_K4115040 BBa_K4115040 ], which is submitted by [https://2022.igem.wiki/shanghaitech-china/ ShanghaiTech_China 2022].<br><br> | ||
| + | |||
<!-- DON'T DELETE --><partinfo>BBa_C0161 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_C0161 EndReviews</partinfo> | ||
Latest revision as of 08:12, 12 October 2022
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0161
Characterization of BBa_C0161-Arizona_State 2016
Authors: Ernesto Luna, Brady Dennison, Cassandra Barrett, Jimmy Xu, Jiaqi Wu, Dr. Karmella Haynes
Our team helped increase characterization of the part BBa_C0161 (LuxI). This part was tested against its ability to induce the part BBa_F2620 by the Canton Lab(MIT). This part outputs PoPS as a Receiver Device combined with LuxR. An induction test on BBa_F2620 had been done by Dr. Barry Canton (2008), but they tested GFP production over various AHL concentrations, while our test was an 8-hour GFP read over time for 2 AHL concentrations (10 and 50%). In addition, the Canton test utilized synthetic 3-O-C6-HSLs while our test utilized AHLs produced via an E.coli chassis. A visual induction test was also done, plating the Sender alongside a GFP positive control, negative receiver control, and F2620.
As shown below, Lux is able to induce F2620, as some colonies in the top right section began producing GFP. This is the expected result, since the Receiver of this system naturally accepts 3-O-C6-HSLs.
The figure below compares LuxI at 10% and 50% concentrations. LuxI is shown to induce F2620. This affirms that F2620 is capable of being induced by LuxI synthesized within BL21(DE3) E. coli.
AHL Disposal Test
The final experiment conducted using this part aimed to determine proper safe disposal procedures for the 3-O-C6-HSL. This AHL molecule is capable of crosstalk with potentially pathogenic strains of bacteria, and proper disposal of these AHLs should be an important biosafety measure taken. S.A. Borchardt had already tested the susceptibility of AHLs to bleach and found that 3-oxo AHLs were easily broken down by bleach while other AHLs were not. Our experiment aimed to test the application of standard EH&S sanitation protocols on AHLs (10% bleach solution and autoclaving). The figure below indicates that AHLs produced by LuxI were not properly deactivated by a 10% bleach solution. This was NOT the expected result, as LuxI produces a 3-oxo AHL, which should have been destroyed by bleach.
A standard 15 minute Liquid autoclave cycle was also used to treat an extracted AHL solution. The figure below indicates that LuxI was nearly completely destroyed via autoclaving. This was the expected result, as the high pressure and temperatures should deactivate any AHL molecules present.
Conclusion
The results demonstrate that Lux was able to effectively induce F2620 after being extracted. The Lux results were not consistent, which showed decreased induction when treated with bleach, but no evidence of complete AHL inactivation. According to the autoclave results, a standard 15 min liquid procedure is able to degrade nearly all AHLs. The extreme pressure and temperature generated by the autoclave was more than enough to remove any threat posed by these AHL samples. In summary, our data suggests that autoclaving will ensure at least some inactivation of AHLs, while bleach may have no effect at all on the degradation of AHLs.
User Reviews
UNIQ1903a1717f90d784-partinfo-00000000-QINU
|
•••••
ETH Zurich 2014 |
Induction by the Supernatant of a LuxI Producing CultureThe production of inducer 3OC6-HSL for the acitivation of the 3OC6-HSL/LuxR dependent promoter pLux (R0062) by LuxI could be shown using the supernatant of a culture containing E. coli cells constitutively expressing LuxI. This supernatant was then used to induce a 3OC6-HSL sensor construct with the reporter sfGFP in another cell culture. Background informationLuxI synthesizes 3OC6-HSL which can bind to the regulator LuxR. The LuxR/3OC6-HSL complex can subsequently induce the promoter pLux. In this experiment we used E. coli TOP10 strain transformed with a plasmid containing a pBBR1 origin of replication and a tetracycline resistance gene, the constitutive promoter J23100 from the Anderson collection, an optimized RBS for this genetic context and the gene for the 3OC6-HSL synthetase LuxI. The RBS was optimized using the [http://salis.psu.edu/software/RBSLibraryCalculatorSearchMode Salis Lab RBS Calculator]. This strain was used as a 3OC6-HSL producer strain. The detailed construct design and full sequence (piG0050max) is [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. As sensor strain we used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter pLux within a [http://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator system] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts BBa_B0040 and BBa_B0015 were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and one of three promoters chosen from the Anderson promoter collection followed by LuxR. A third plasmid containing the pBBR1 origin and a tetracycline resistance was introduced, since the LuxI expressing 3OC6-HSL-producer strain contained a tetracycline resistance an therefore the supernatant also contained tetracycline. The detailed construct designs and full sequences (piG0041, piG0065) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. Experimental SetupThe above described E. coli TOP10 AHL producer strain was grown in Lysogeny Broth (LB) containing tetracycline (10 μg/mL) to an OD600 of 1.2 (37 °C, 220 rpm). The supernatant was harvested by centrifugation at 20'000 g for 5 minutes. Dilutions of 100%, 85%, 70%, 55%, 40%, 25%, 10%, 5%, 1%, and 0% (v/v) supernatant/LB were prepared and kanamycin, ampicillin, and tetracycline were added to a final concentration of 50 μg/mL, 200 μg/mL, and 10 μg/mL respectively. The dilutions were aliquoted in triplicates in microtiter plate format on 96-well plates (200 μL volume). The above described E. coli TOP10 sensor strain was grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL), ampicillin (200 μg/mL), and tetracycline (10 μg/mL) to an OD600 of about 1.5 (37 °C, 220 rpm). As a reference, a preculture of the same strain lacking the sfGFP gene was included for each assay. The cultures were then diluted 1:40 in the prepared supernatant/LB mixtures containing the appropriate antibiotics and measured in triplicates in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h at 37 °C with a Tecan infinite M200 PRO plate reader (optical density measured at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). From the the obtained kinetic data, we calculated mean values and plotted the dose-response-curve for 360 min past inoculation. ResultsIt could be shown that the supernatant of the LuxI producer strain can induce the LuxR/pLux sensor strain. The ON/OFF ratio between 0% and 100% supernatant is approximately 33. The increasing fluorescence per OD600 with increasing proportion of supernatant is shown in Figure 1.  Figure 1: Dose-response curve with supernatant of a LuxI producer culture. The supernatant of E. coli cells gown in LB medium, constitutively expressing LuxI, was used in different dilutions with fresh LB medium as growth medium for E. coli cells containing a construct expressing sfGFP under thr pLux promoter and constitutively expressing the regulator LuxR. Fluorescence/OD600 of sfGFP was measured 6 h after inoculation. Measurements were conducted in triplicates. The squares depict the mean values and the bars represent +/- the standard deviation of the measured triplicates.
|
|
Antiquity |
This review comes from the old result system and indicates that this part did not work in some test. |
ShanghaiTech_China 2022's Characterization
Design
LuxI can send AHL, 3OC6 HSL here and the signal can be received by the LuxR(BBa_C0062). LuxR can then regulate the downstream Lux pR (BBa_R0062) to make superfolder GFP (BBa_K4115000) express.(Figure 1)
Results
We use LuxI supernatant to induce LuxR to respond and express sfGFP.The test group is compared with 10-7M 3OC6 HSL induced group and no induced group(blank group). The result is shown by dividing the fluorescence intensity by the OD600, both measured in the transformed E. coil. (Figure 2)
We also induced the expression of LuxR in sodium alginate gel beads and detected the expression of sfGFP under UV light. (Figure 3)
More detailed information can be seen on the composite page BBa_K4115040 , which is submitted by ShanghaiTech_China 2022.
UNIQ1903a1717f90d784-partinfo-00000003-QINU