Difference between revisions of "Part:BBa K1499004"
(→Verification of Part) |
|||
| (17 intermediate revisions by 6 users not shown) | |||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
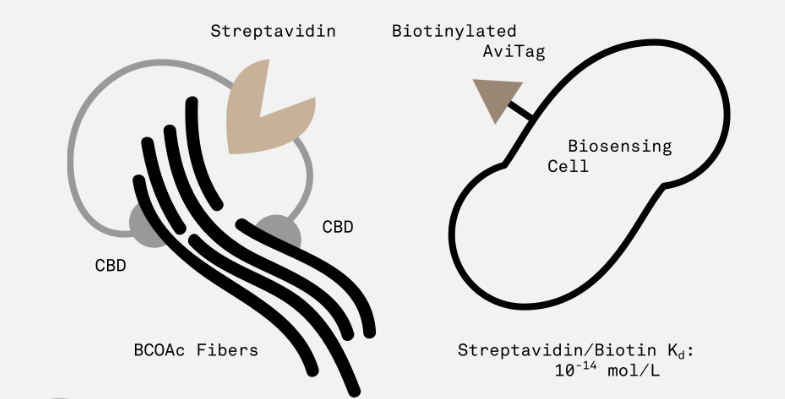
| − | The idea behind this part is that each domain binds to a single cellulose fiber, thus providing a way to cross-link and strengthen cellulose polymers. In addition, the streptavidin domain allows for the modular addition of biotinylated | + | The idea behind this part is that each domain binds to a single cellulose fiber, thus providing a way to cross-link and strengthen cellulose polymers. In addition, the streptavidin domain allows for the modular addition of sensor cells with a biotinylated AviTag peptide (expressed on the surface of the cell by appending it to outer membrane protein OmpA). |
| + | |||
| + | [[Image:Cross_linker_desc.png|750px|thumb|center|<b>Figure 1.</b> A schematic of CBD fusion protein and sensor cell surface protein complex interaction.]] | ||
<!-- --> | <!-- --> | ||
| Line 15: | Line 17: | ||
===Verification of Part=== | ===Verification of Part=== | ||
| − | The part was sequence verified before submission to the registry with two reads using VF2 and VR (Figure 1). | + | The part was sequence verified before submission to the registry with two reads using VF2 and VR. |
| + | |||
| + | [[Image:CBD_Forward_Sequence.png|750px|thumb|center|<b>Figure 2.</b> The forward read using VF2.]] | ||
| + | |||
| + | [[Image:CBD_Reverse_Sequence.png|750px|thumb|center|<b>Figure 3.</b> The reverse read using VR.]] | ||
| + | |||
| + | ===Results=== | ||
| + | We plan to assay the functionality of this part using a GFP construct inside a cell with the AviTag/OmpA surface complex. We will purify and apply our CBD/Streptavidin fusion protein to a cellulosic surface, followed by the above transformed cells, and attempt to wash off the cells using an isotonic solution. If the level of fluorescence does not change, we can assume our system has been successful. | ||
| + | |||
| + | The 2016 Stanford-Brown iGEM Team purified this linker protein and used it to create a BioDevice. Used in tandem with a biotinylated fluorophore, this CBD/Streptavidin fusion protein served as a linker between cellulose paper and the fluorophore-quencher biosensor described here: http://2016.igem.org/Team:Stanford-Brown/SB16_BioSensor_FQsensor. | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | |width='60%' valign='top'| | ||
| + | <h2> INSA Lyon 2016 Experiments on this part </h2> | ||
| + | |||
| + | =='''Characterization'''== | ||
| + | <html> | ||
| + | <h3 id="CBD">1. Purification Using Cellulose Affinity</h3> | ||
| + | <p>The <a href="https://parts.igem.org/Part:BBa_K1934020">BBa_K1934020</a> part conceived by the 2016 INSA-Lyon team and synthesized by IDT was cloned into pSB1C3 and transformed into the <i>E. coli</i> NM522 strain. One recombinant clone was grown overnight in LB at 24°C, with IPTG 1 mmol.L<sup>-1</sup> and glucose 5 mmol.L<sup>-1</sup>. Cells were harvested and resuspended in 1 mL lysis buffer (50 mmol.L<sup>-1</sup> Tris, 300 mmol.L<sup>-1</sup> NaCl, 10% glycerol). Then the mix was sonicated 5 times 30 seconds on ice at moderate power. The lysate was centrifuged at 14,000 g for 10 min. The supernatant was treated as follow: | ||
| + | <ul style="list-style-type:circle"> | ||
| + | <li>Wash microcrystaline Cellulose five times in water. Then equilibrate in washing buffer (ammonium sulfate 1M). Pack the cellulose (10x10mm) in small chromatography columns (we used syringes barrels).</li> | ||
| + | <li>Gently pour the lysate supernatant on the column. Once the liquid starts flowing through evenly, measure the OD<sub>280</sub> of the different fractions. Continue pouring washing buffer until the OD<sub>280</sub> stabilizes around zero.</li> | ||
| + | <li>Change the washing buffer to water. OD<sub>280</sub> shortly rises. Keep the fractions with the highest OD<sub>280</sub> . They should contain the protein.</li> | ||
| + | <li>Analyse collected fractions on an SDS-PAGE. | ||
| + | Optionally, proteins may be concentrated using ultrafiltration.</li> | ||
| + | </ul></p> | ||
| + | |||
| + | |||
| + | |||
| + | <figure><p style="text-align:center;"><img src="https://static.igem.org/mediawiki/2016/8/8b/INSA-Lyon_SCBD_elution.jpeg" width = "800"/><figcaption><b>Figure 1. Purification of the chimeric Streptavidin-CBD protein on a cellulose column</b> This elution graph shows a first peak, present for both the control and our expression culture. This first peak corresponds to unbound proteins. In the presence of water, only one peak was observed: it’s the elution peak of our protein.</figcaption></figure> | ||
| + | |||
| + | <h3 id="RT">2. <a href="https://parts.igem.org/Part:BBa_K1934020">BBa_K1934020</a> encodes a protein able to bind both biotin and cellulose</h3> | ||
| + | <figure><p style="text-align:center;"><img src= "https://static.igem.org/mediawiki/2016/f/f2/INSA-Lyon_principle_SCBD.jpeg" width = "400"/><figcaption> | ||
| + | |||
| + | Affinity of the streptavidin-CBD encoded by <a href="https://parts.igem.org/Part:BBa_K1934020">BBa_K1934020</a> to cellulose was compared to the one of commercial streptavidin. A molecule of fluorescein was grafted at the 5’ end of a DNA oligo carrying a molecule of biotin at its 3’ end. This DNA oligo constitutes the reporter system. Such modified oligo was mixed either with the engineered <a href="https://parts.igem.org/Part:BBa_K1934020">streptavidin-CBD</a> or with commercial streptavidin. The resulting mix was incubated with microcrystalline cellulose in presence of PBS for 1 hour. The cellulose was then washed twice with fresh PBS and fluorescence was measured. Every experiment was done in triplicate. | ||
| + | |||
| + | |||
| + | <figure><p style="text-align:center;"><img src= "https://static.igem.org/mediawiki/2016/f/fa/INSA-Lyon_SCBD_linking.jpeg" width = "400"/><figcaption><b>Figure 2. The Streptavidin-CBD is able to bind biotin and cellulose.</b> Mixed raw cellulose with our report system shows no fluorescence (first bar). The measured fluorescence indicates that commercial streptavidin was able to bind our reporter system and sticks at a low extent to cellulose. We concluded that this results from none-specific adsorption. For the streptavidin-CBDs part <a href="https://parts.igem.org/Part:BBa_K1934020">(BBa_K1934020)</a>, a high fluorescent signal was recorded. </figcaption></figure> | ||
| + | <strong>This experiment shows that this streptavidin-CBD protein is able to bind efficiently biotin and cellulose at the same time. </strong> | ||
| + | The same experiment was done for the <a href="https://parts.igem.org/Part:BBa_K1934030">BBa_K1934030</a>: part displaying a different cellulose binding domain, namely CBD-CipA. The binding efficiency of streptavidin-CBDs tends to be slightly lower compared to streptavidin-CipA (x1.1) but was not statistically demonstrated. | ||
| + | |||
| + | |||
| + | <h3 id="RT">3. Streptavidin-CBDs modeling</h3> | ||
| + | |||
| + | <figure><p style="text-align:center;"><img src= "https://static.igem.org/mediawiki/2016/0/06/T--INSA-Lyon--Streptavidin_CBDs.png" width = "400"/><figcaption> | ||
| + | |||
| + | We made a homology modeling as a confirmation of the working protein folding. The domains didn't seem well defined because of the hindered, but we still conserved the secondary structure of the protein. The use of a linker may be appropriate to allow a better efficiency. | ||
| + | |||
| + | </html> | ||
| + | |} | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1499004 AddReview 4</partinfo> | ||
| + | <I>mbecich</I> | ||
| + | |width='60%' valign='top'| | ||
| + | The part worked as specified for our needs! We cloned it in pSB1C3, with a pTac promotor, strong RBS and a double terminator. It is important to note that this protein tends to dimerize when it is over-expressed driving to a loss of function. | ||
| + | |} | ||
| + | ==Improvements by SHSBNU_China 2021== | ||
| + | BBa_K3798033 https://parts.igem.org/Part:BBa_K3798033 is modified from this part. From BBa_K1499004, we optimized the Condon usage of this protein for Bacillus subtilis. CBD-SA is the linker module which connects cellulose and aptamer. It is a fusion protein composed of two cellulose-binding-domain and one Streptavidin domain that is produced by engineered Bacillus subtilis. The actual structure of this linker module refers to CBD-SA-CBD. We wanted to use system with B. subtilis to optimize our production of CBD-SA. But, the plasmids of CBD-SA were not transformed successfully into competent cells of B. subtilis. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1499004 parameters</partinfo> | <partinfo>BBa_K1499004 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 12:27, 21 October 2021
Cellulose binding domains with streptavidin domain generator
This part encodes a protein with two cellulose-binding domains on either end joined by a streptavidin domain.
Usage and Biology
The idea behind this part is that each domain binds to a single cellulose fiber, thus providing a way to cross-link and strengthen cellulose polymers. In addition, the streptavidin domain allows for the modular addition of sensor cells with a biotinylated AviTag peptide (expressed on the surface of the cell by appending it to outer membrane protein OmpA).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 703
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 304
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 651
Characterization
Verification of Part
The part was sequence verified before submission to the registry with two reads using VF2 and VR.
Results
We plan to assay the functionality of this part using a GFP construct inside a cell with the AviTag/OmpA surface complex. We will purify and apply our CBD/Streptavidin fusion protein to a cellulosic surface, followed by the above transformed cells, and attempt to wash off the cells using an isotonic solution. If the level of fluorescence does not change, we can assume our system has been successful.
The 2016 Stanford-Brown iGEM Team purified this linker protein and used it to create a BioDevice. Used in tandem with a biotinylated fluorophore, this CBD/Streptavidin fusion protein served as a linker between cellulose paper and the fluorophore-quencher biosensor described here: http://2016.igem.org/Team:Stanford-Brown/SB16_BioSensor_FQsensor.
INSA Lyon 2016 Experiments on this partCharacterization
1. Purification Using Cellulose AffinityThe BBa_K1934020 part conceived by the 2016 INSA-Lyon team and synthesized by IDT was cloned into pSB1C3 and transformed into the E. coli NM522 strain. One recombinant clone was grown overnight in LB at 24°C, with IPTG 1 mmol.L-1 and glucose 5 mmol.L-1. Cells were harvested and resuspended in 1 mL lysis buffer (50 mmol.L-1 Tris, 300 mmol.L-1 NaCl, 10% glycerol). Then the mix was sonicated 5 times 30 seconds on ice at moderate power. The lysate was centrifuged at 14,000 g for 10 min. The supernatant was treated as follow:
2. BBa_K1934020 encodes a protein able to bind both biotin and cellulose
|
|
••••
mbecich |
The part worked as specified for our needs! We cloned it in pSB1C3, with a pTac promotor, strong RBS and a double terminator. It is important to note that this protein tends to dimerize when it is over-expressed driving to a loss of function. |
Improvements by SHSBNU_China 2021
BBa_K3798033 https://parts.igem.org/Part:BBa_K3798033 is modified from this part. From BBa_K1499004, we optimized the Condon usage of this protein for Bacillus subtilis. CBD-SA is the linker module which connects cellulose and aptamer. It is a fusion protein composed of two cellulose-binding-domain and one Streptavidin domain that is produced by engineered Bacillus subtilis. The actual structure of this linker module refers to CBD-SA-CBD. We wanted to use system with B. subtilis to optimize our production of CBD-SA. But, the plasmids of CBD-SA were not transformed successfully into competent cells of B. subtilis.