Difference between revisions of "PBAD SPL"
(→Characterization) |
(→The library) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 17: | Line 17: | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 02 |
|<font face="Courier" size="+1" class=">CTGACGACGCCCCTCTCCGCCCCTTAAAATGTCCA</font> | |<font face="Courier" size="+1" class=">CTGACGACGCCCCTCTCCGCCCCTTAAAATGTCCA</font> | ||
|align="center"|14.2 | |align="center"|14.2 | ||
| Line 23: | Line 23: | ||
|align="center"|69.9 | |align="center"|69.9 | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 03 |
|<font face="Courier" size="+1">CTGACGTAATCATACCGCCGAAAGTATTATCATTA</font> | |<font face="Courier" size="+1">CTGACGTAATCATACCGCCGAAAGTATTATCATTA</font> | ||
|align="center"|16.6 | |align="center"|16.6 | ||
| Line 29: | Line 29: | ||
|align="center"|39 | |align="center"|39 | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 04 |
|<font face="Courier" size="+1">CTGACGCGACAACATTGCGTCCTATAAAATGCCGA</font> | |<font face="Courier" size="+1">CTGACGCGACAACATTGCGTCCTATAAAATGCCGA</font> | ||
|align="center"|14.2 | |align="center"|14.2 | ||
| Line 35: | Line 35: | ||
|align="center"|41 | |align="center"|41 | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 05 |
|<font face="Courier" size="+1">CTGACGGCCCGCCCCCGATGCGCATAAAATACCAA</font> | |<font face="Courier" size="+1">CTGACGGCCCGCCCCCGATGCGCATAAAATACCAA</font> | ||
|align="center"|17.7 | |align="center"|17.7 | ||
| Line 53: | Line 53: | ||
|align="center"|38.8 | |align="center"|38.8 | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 09 |
|<font face="Courier" size="+1">CTGACGCCACCCCCCCCCCCGGCGTATAATTCCCA</font> | |<font face="Courier" size="+1">CTGACGCCACCCCCCCCCCCGGCGTATAATTCCCA</font> | ||
|align="center"|16.1 | |align="center"|16.1 | ||
| Line 59: | Line 59: | ||
|align="center"|30.4 | |align="center"|30.4 | ||
|- | |- | ||
| − | |align="center"|Col. | + | |align="center"|Col. 08 |
|<font face="Courier" size="+1">CTGACGCCCTACCGCTCGCCCCCCTATTATACCCA</font> | |<font face="Courier" size="+1">CTGACGCCCTACCGCTCGCCCCCCTATTATACCCA</font> | ||
|align="center"|13.4 | |align="center"|13.4 | ||
| Line 145: | Line 145: | ||
var letter = amino_string[j]; | var letter = amino_string[j]; | ||
var color = "black"; | var color = "black"; | ||
| + | //if( letter == "C"){ | ||
| + | // color = "green"; | ||
| + | //} else if( letter == "A"){ | ||
| + | // color = "blue"; | ||
| + | //} else if( letter == "T"){ | ||
| + | // color = "orange"; | ||
| + | //} else if( letter == "G"){ | ||
| + | // color = "red"; | ||
| + | //} | ||
| + | //$(amino_string_container).append("<font color=" + color + ">" + letter + "</font>"); | ||
if( letter == "C"){ | if( letter == "C"){ | ||
| − | color = " | + | color = "rgb(26,204,26)"; |
} else if( letter == "A"){ | } else if( letter == "A"){ | ||
| − | color = " | + | color = "rgb(128,179,230)"; |
} else if( letter == "T"){ | } else if( letter == "T"){ | ||
| − | color = " | + | color = "rgb(230,153,77)"; |
} else if( letter == "G"){ | } else if( letter == "G"){ | ||
| − | color = " | + | color = "rgb(230,128,128)"; |
} | } | ||
| − | $(amino_string_container).append( | + | $(amino_string_container).append('<font style="background-color: ' + color + '">' + letter + '</font>'); |
} | } | ||
} | } | ||
| Line 167: | Line 177: | ||
=Characterization= | =Characterization= | ||
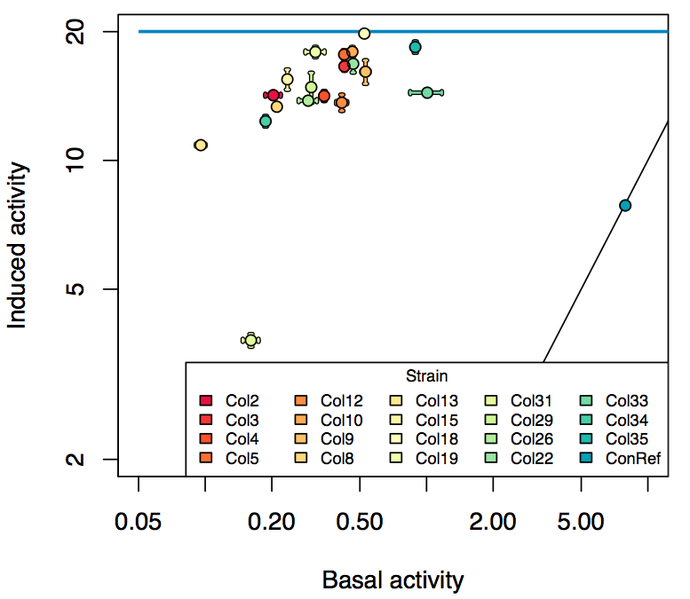
| − | We did a characterization by running growth experiments in parallel on a BioLector ( | + | We did a characterization by running growth experiments in parallel on a BioLector ([http://2013.igem.org/Team:DTU-Denmark/pBAD_SPL see more]). Below diagram sums up our results: |
| − | [File:690px-Induced vs basal.png| | + | [[File:690px-Induced vs basal.png|600px|center|Diagram over the promoter strength]] |
=Conclusion= | =Conclusion= | ||
| + | The pBAD SPL contain promoters to the arabinose inducible system that will enable low leakiness but still maintain a reasonable promoter strength when turned on. This verifies that the SPL standard ([http://dspace.mit.edu/handle/1721.1/60080 RFC 63]) also can be used to produce promoters for inducible systems. | ||
Latest revision as of 22:06, 4 October 2013
Description
This is a page containing a synthetic promoter library (SPL) of pBAD promoters made by [http://2013.igem.org/Team:DTU-Denmark 2013 DTU iGEM team]. The library can be used to make and arabinose inducible system with very low leakiness, or tune the the promoter strength of the system when it's turned on.
A tight inducible system can be a very useful tool to express lethal or growth inhibiting proteins in E.coli. It can also be necessary with a non leaky expression system if you would like to visualize the expression of GFP SF in E.coli (see [http://2013.igem.org/Team:DTU-Denmark/HelloWorld link]).
The library
Here is a complete list of the sequences and their relative leakiness and strength:
| Identifier | Sequencea | Induced Strength | Leakiness | Inducibilityb | ||
|---|---|---|---|---|---|---|
| Col. 02 | CTGACGACGCCCCTCTCCGCCCCTTAAAATGTCCA | 14.2 | 0.203 | 69.9 | ||
| Col. 03 | CTGACGTAATCATACCGCCGAAAGTATTATCATTA | 16.6 | 0.426 | 39 | ||
| Col. 04 | CTGACGCGACAACATTGCGTCCTATAAAATGCCGA | 14.2 | 0.345 | 41 | ||
| Col. 05 | CTGACGGCCCGCCCCCGATGCGCATAAAATACCAA | 17.7 | 0.424 | 41.6 | ||
| Col. 12 | CTGACGCGTATCGCGAGCGGGCGTTATTATACGCA | 13.6 | 0.414 | 32.9 | ||
| Col. 10 | CTGACGCCCCGAATCAGTAGTATTTATTATCTAGA | 18 | 0.462 | 38.8 | ||
| Col. 09 | CTGACGCCACCCCCCCCCCCGGCGTATAATTCCCA | 16.1 | 0.530 | 30.4 | ||
| Col. 08 | CTGACGCCCTACCGCTCGCCCCCCTATTATACCCA | 13.4 | 0.211 | 63.4 | ||
| Col. 13 | CTGACGCCCCCCCCGTCCACCCCCTAATATCCCGA | 10.9 | 0.096 | 113.6 | ||
| Col. 15 | CTGACGTGCGCCTGCCGTCCAAAGTAATATCCTTA | 15.5 | 0.235 | 65.8 | ||
| Col. 18 | CTGACGCCCCGAATCAGTAGTATTTATTATCTAGA | 19.8 | 0.524 | 37.8 | ||
| Col. 19 | CTGACGCTCGCACCGGCGACCCGATAATATCCATA | 18.0 | 0.315 | 57 | ||
| Col. 31 | CTGACGCCCCCCCTCCCCCGACCTAAAATATCCAG | 3.8 | 0.161 | 23.6 | ||
| Col. 29 | CTGACGCCGCTGCACCCGTCCCCCTATTATAGCAA | 14.8 | 0.301 | 49.2 | ||
| Col. 26 | CTGACGCCGATCCTATCTCCCTATTAAAATTTTTA | 13.8 | 0.292 | 47.2 | ||
| Col. 22 | CTGACGCCCCGAATCAGTAGTATTTATTATCTAGA | 16.8 | 0.466 | 36.1 | ||
| Col. 33 | CTGACGCCAGCTTCCTACTTCAAATATAATCCGTA | 14.4 | 1.01 | 14.3 | ||
| Col. 34 | CTGACGTCCCTCTTCCGCGCCCCCTAAAATACCCA | 12.4 | 0.187 | 65.9 | ||
| Col. 35 | CTGACGCCCAACCCGACACAGCGATATAATAATGA | 18.4 | 0.89 | 20.7 | ||
a The sequence of each pBAD promoter characterized. To use this simply produce a primer with the promoter sequence you wish in a tail. Replace the old promoter in your system e.g. Part:BBa_K808000 with the new and see the chance. For more information go to ([http://2013.igem.org/Team:DTU-Denmark/pBAD_SPL link]).
b Inducibility = Induced Strength divided by leakiness.
Characterization
We did a characterization by running growth experiments in parallel on a BioLector ([http://2013.igem.org/Team:DTU-Denmark/pBAD_SPL see more]). Below diagram sums up our results:
Conclusion
The pBAD SPL contain promoters to the arabinose inducible system that will enable low leakiness but still maintain a reasonable promoter strength when turned on. This verifies that the SPL standard ([http://dspace.mit.edu/handle/1721.1/60080 RFC 63]) also can be used to produce promoters for inducible systems.