Difference between revisions of "Part:BBa K592009:Experience"
MikeFerguson (Talk | contribs) |
MatildaBrink (Talk | contribs) |
||
| (42 intermediate revisions by 13 users not shown) | |||
| Line 4: | Line 4: | ||
===Applications of BBa_K592009=== | ===Applications of BBa_K592009=== | ||
| + | ===User review of SHSBNU_China=== | ||
| + | [[File: SHSBNU 17 40aL1.jpg|600px|thumb|center|Figure 1]] | ||
| + | |||
| + | Figure 1 TtrS/R system and ThsS/R system | ||
| + | ThsS (BBa_K2507000) and ThsR (BBa_K2507001), both codon-optimized for E. coli, are two basic parts which belong to the two-component system from the marine bacterium Shewanella halifaxensis. | ||
| + | By linking thsR with chromoprotein genes (BBa_K2507009, BBa_K2507010, BBa_K2507011) or the violacein producing operon vioABDE (BBa_K2507012), this system can respond to thiosulfate by producing a signal visible to the naked eye, such as chromoproteins (spisPink-pink chromoprotein, gfasPurple-purple chromoprotein, amilCP-blue chromoprotein) or a dark-green small-molecule pigment (protoviolaceinic acid). | ||
| + | E. coli-codon-optimized TtrS(BBa_K2507002) and TtrR (BBa_K2507003) are two basic parts which are derived from the two-component system of the marine bacterium Shewanella baltica. | ||
| + | |||
| + | By linking ttrR with chromoprotein genes (BBa_K2507009, BBa_K2507010, BBa_K2507011) or the violacein producing operon vioABDE (BBa_K2507012), this system can respond to thiosulfate by producing a signal visible to the naked eye such as chromoproteins (spisPink-pink chromoprotein, gfasPurple-purple chromoprotein, amilCP-blue chromoprotein) or a dark-green small-molecule pigment (protoviolaceinic acid). | ||
| + | |||
| + | [[File: SHSBNU 17 40aL2.jpg|600px|thumb|center|Figure 2]] | ||
| + | |||
| + | Figure 2. Characterization of the ThsS/R and TtrS/R system by observing the chromprotein expression levels. We added 1mM, 0.1mM, 0.01mM and 0 Na2S2O3 to ThsS/R system and added 2.5mM, 1mM, 0.1mM and 0 Na2S4O6·2H2O to ThsS/R system. The results demonstrate there is an obvious response in the ThsS/R system. In a. gfasPurple system, the response curve is obvious, and b. spisPink & c. amilCP with rather heavy leaky expression without inducer. d.e.f. However, in TtrS/R system there is no clear result. | ||
| + | |||
| + | [[File: SHSBNU 17 40aL3.jpg|600px|thumb|center|Figure 3]] | ||
| + | |||
| + | Figure 3. | ||
| + | Characterization of the ThsS/R and TtrS/R system by observing the protoviocaceinic acid( dark-green pigment). We added 1mM, 0.1mM, 0.01mM and 0 Na2S2O3 to ThsS/R system and added 2.5mM, 1mM, 0.1mM and 0 Na2S4O6·2H2O to ThsS/R system. The results demonstrate there is an obvious response in the ThsS/R system with rather heavy leaky expression without inducer. And the Ths/R+protoviolaceinic acid works rather well! | ||
| + | |||
| + | |||
| + | |||
| + | |||
===User Reviews=== | ===User Reviews=== | ||
| + | |||
| + | <!-- DON'T DELETE --><partinfo>BBa_K1155003 StartReviews</partinfo> | ||
| + | <!-- Template for a user review | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1155003 AddReview number</partinfo> | ||
| + | <I>Team</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | |}; | ||
| + | <!-- End of the user review template --> | ||
| + | |||
| + | |||
| + | <!-- DON'T DELETE --><partinfo>BBa_K1155003 StartReviews</partinfo> | ||
| + | <!-- Template for a user review | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1155003 AddReview number</partinfo> | ||
| + | <I>Team</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |}; | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <partinfo>BBa_K1155003 EndReviews</partinfo> | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1155003 AddReview 1</partinfo> | ||
| + | <I>Pittsburgh 2016</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | We planned to use this part as a reporter in our circuit. We cloned the sample provided in the 2016 DNA Distribution Kit to a T7 promoter (Part BBa_K525998) and terminator (Part BBa_B0015) but did not see expression in T7 cells or in T7 cell extract. Sequencing results suggested that the DNA sequence was CFP, not amilCP. | ||
| + | |||
| + | |}; | ||
| + | |||
| + | |||
| + | |||
| + | <!-- DON'T DELETE --><partinfo>BBa_K1155003 EndReviews</partinfo> | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K592009 AddReview 5</partinfo> | ||
| + | <I>iGEM Sydney Australia</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | |||
| + | The Sydney_ Australia 2016 team improved this part via error prone PCR for inclusion in our ethylene biosensor. We managed to produce three new colour strains, a purple, a pink, and a mint green. | ||
| + | <br> | ||
| + | |||
| + | [[File: Sydney_australia_amilcp_mutants.png |620px|center]] | ||
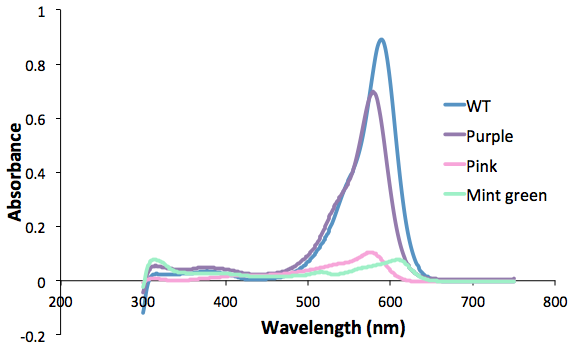
| + | <b>Figure 1. Purple, pink and green mutants compared to the wild type.<br> | ||
| + | Protein was extracted from 50mL of culture and resuspended in 0.5mL TE buffer.</b><br> | ||
| + | [[File:Sydney_australia_amilcp_spectrum.png|620px|center]] | ||
| + | <br> | ||
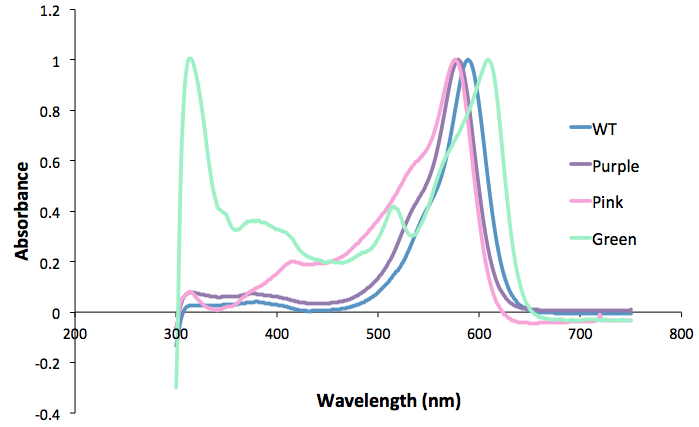
| + | <b>Figure 2. Spectrum scan of the extracted protein from the different coloured amilCP mutants. Protein concentration is equal between all samples.</b><br> | ||
| + | [[Image:Sydney_australia_amilcp_spectrum4.png |620px|center]] | ||
| + | <br> | ||
| + | <b>Figure 3. Spectrum scan of the extracted protein from the different coloured amilCP mutants, with the spectrums all adjusted to the same levels to show peaks.</b> | ||
| + | |}; | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K592009 AddReview 5</partinfo> | ||
| + | <I>iGEM Uppsala 2013</I> | ||
| + | |width='60%' valign='top'| | ||
| + | [[Image:High_and_low_temp_amilCP.jpg|620px|center]] | ||
| + | |||
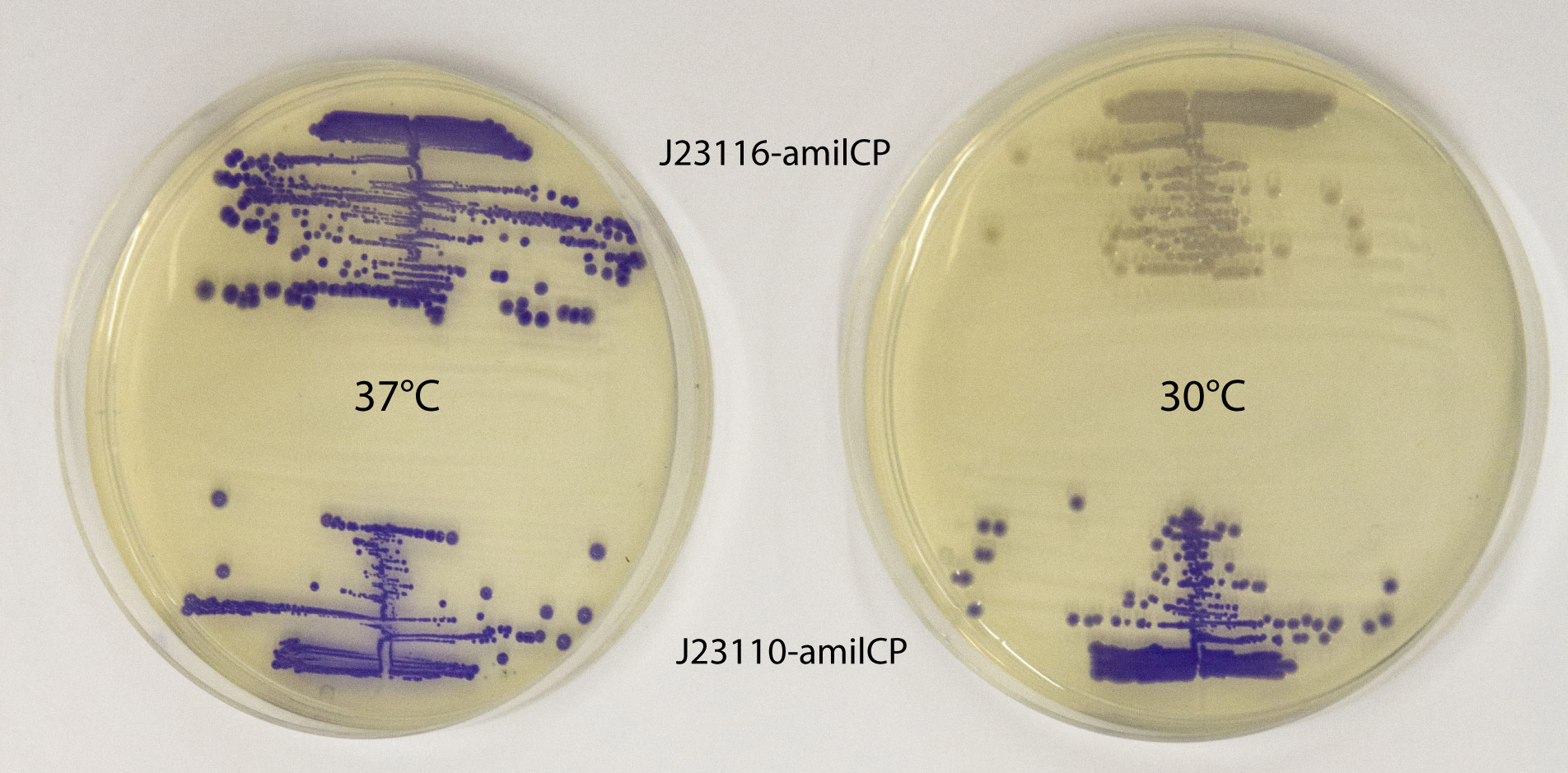
| + | This part works well as a reporter. As expected, different levels of expression of amilCP in E. coli MG1655 generates different intensities in color. amilCP expressed from the high copy plasmid pSB1C3 by weak promoter J23116 (top part of plates) and strong promoter J23110 (bottom part of plates) at 37 °C (left plate) and 30 °C (right plate) respectively. After incubation at 30 °C a clear difference in color intensity can be seen between the expression from the two different promoters, while after incubation at 37 °C the difference in intensity is much less pronounced. This indicates that expression from the promoters are temperature dependent, and it shows that the quantifiable range of the chromoproteins is somewhat limited. Plates were incubated for 23 h at 37 °C or 30 °C followed by 20 h at 22 °C. | ||
| + | |}; | ||
| + | |||
| + | <partinfo>BBa_K1155003 StartReviews</partinfo> | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K1155003 AddReview 5</partinfo> | ||
| + | <I>Paris_Saclay 2013</I> | ||
| + | |width='60%' valign='top'| | ||
| + | Our team improved the biobrick BBa_K592009 by assembly of biobricks B0034, BBa_K592009 and B0015 to construct a composite part including a ribosome binding site, full length of amilCP gene and double terminator. We used it as one of our reporter gene in Escherichia coli to controlled activator or repressor activity of our promotor under aerobic or anaerobic conditions (Pndh*, PnirB, PnarG, PnarK, PbphR1, PbphR2). | ||
| + | |||
| + | Construction : Pndh* (repressor) + BBa_K1155003 : | ||
| + | * In anaerobic conditions : Pndh* is repressed --> BBa_K1155003 isn't expessed --> no color | ||
| + | * In aerobic conditions : Pndh* isn't repressed --> BBa_K1155003 is expressed --> violet color | ||
| + | |||
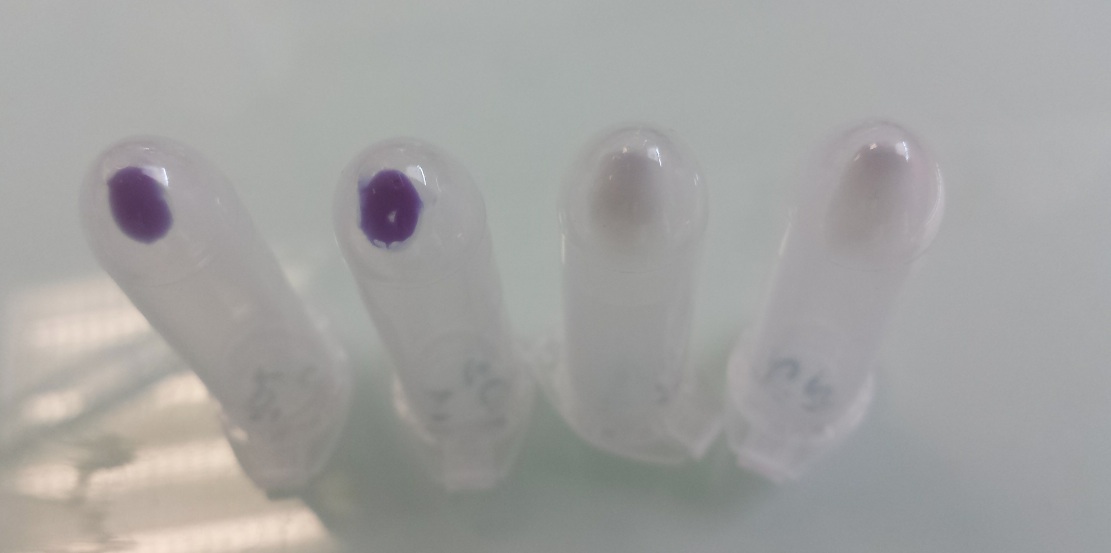
| + | [[File:PsPfnr3008.jpg|500px]] | ||
| + | |||
| + | Tubes 1 and 2 : cellular pallet of cultures grown in aerobic conditions | ||
| + | |||
| + | Tubes 3 and 4 : cellular pallet of cultures grown in anaerobic conditions | ||
| + | |}; | ||
| + | <partinfo>BBa_K1155003 EndReviews</partinfo> | ||
| + | |||
| + | |||
<!-- DON'T DELETE --><partinfo>BBa_K592009 StartReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K592009 StartReviews</partinfo> | ||
<!-- Template for a user review | <!-- Template for a user review | ||
| Line 11: | Line 140: | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | <partinfo>BBa_K592009 AddReview | + | <partinfo>BBa_K592009 AddReview 5</partinfo> |
<I>iGEM Groningen 2012</I> | <I>iGEM Groningen 2012</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
|}; | |}; | ||
| + | |||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_K592009 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K592009 EndReviews</partinfo> | ||
| Line 22: | Line 152: | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | |||
| − | |||
<partinfo>BBa_K592009 AddReview 5</partinfo> | <partinfo>BBa_K592009 AddReview 5</partinfo> | ||
| + | <I>iGEM Michigan 2013</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
The 2013 Michigan igem team successfully used this part to produce amilCP. | The 2013 Michigan igem team successfully used this part to produce amilCP. | ||
| Line 32: | Line 163: | ||
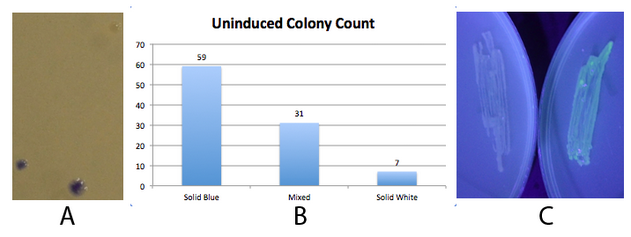
NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON. | NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON. | ||
| + | |}; | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | |||
<partinfo>BBa_E1010 AddReview 5</partinfo> | <partinfo>BBa_E1010 AddReview 5</partinfo> | ||
| Line 69: | Line 206: | ||
After induction of the promoter before AmilCP, B. subtilis also turned clearly purple/blue. Please have a look at the page of [[Part:BBa_K818400|BBa_K818400]] for more information. | After induction of the promoter before AmilCP, B. subtilis also turned clearly purple/blue. Please have a look at the page of [[Part:BBa_K818400|BBa_K818400]] for more information. | ||
| + | |}; | ||
| + | {|width='80%' style='border:1px solid gray' | ||
|- | |- | ||
|width='10%'| | |width='10%'| | ||
| − | |||
<partinfo>BBa_K592009 AddReview number</partinfo> | <partinfo>BBa_K592009 AddReview number</partinfo> | ||
| Line 80: | Line 218: | ||
'''iGEM Team Uppsala University 2012''' | '''iGEM Team Uppsala University 2012''' | ||
| − | We have observed that expression of amilCP confers a noticeable fitness cost in E coli. This is surprising, given that such behaviour is not seen in other homologous chromoproteins and fluorescent proteins. We thus recommend using the new aeBlue (<partinfo>K864401</partinfo>) chromoprotein for blue color expression, which also has a more clear blue color. | + | We have observed that expression of amilCP confers a noticeable fitness cost in E coli. This is surprising, given that such behaviour is not seen in other homologous chromoproteins and fluorescent proteins. We thus recommend using the new aeBlue (<partinfo>K864401</partinfo>) chromoprotein for blue color expression, which also has a more clear blue color. !!!!!!! |
| + | |}; | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K592009</partinfo> | ||
| + | <I>iGEM DTU-Denmark 2017</I> | ||
| + | |width='60%' valign='top'| | ||
| + | |||
| + | The DTU-Denmark 2017 team has added a 6 x his-tag to the C-terminal end of the amilCP protein, to ensure easy purification of amilCP using affinity chromatography. The his-tagged amilCP was submitted to the parts registry as [[Part:BBa_K2355002|BBa_K2355002]]. | ||
| + | |||
| + | The recombinant amilCP with his-tag was expressed in E.coli, and the and the His-tagged amilCP was purified using a Ni-NTA spin colomn. Compared to a control with expressed amilCP from BBa_K592009, the his-tagged amilCP had a 2-4 fold increase in amilCP concentration following the his-tag purification. | ||
| + | Take a look at the entry of [[Part:BBa_K2355002|BBa_K2355002]] for more information. | ||
|}; | |}; | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | <h1>BNU-China 2017 Experience</h1> | ||
| + | <html> | ||
| + | <p>Team UCLA in 2015 iGEM used honeybee silk protein 3 (part number: <a href='https://parts.igem.org/Part:BBa_K1763000'>BBa_K1763000</a>) in their project, which can serve the purpose of being the substitution of spider thread and natural silk. Although it is not as strong as other kind of silk protein, the honeybee silk protein has some advantages compared to spider thread and natural silk: the gene of honeybee silk protein is smaller, with no duplicated sequences, and easier to be synthesized and modified by molecular biology technology. </p> | ||
| + | <p>However, the silk protein produced from UCLA has no color, thus with less aesthetic characteristic and uneasy to observe directly. In order to observe the produced honeybee silk protein more directly, we add a blue chromoprotein, amilCP (part number: <a href='https://parts.igem.org/Part:BBa_K592009'>BBa_K592009</a>) , to the end of the silk protein. AmilCP is a blue chromoprotein comes from coral, which has a strong blue after expressing. The color has the biggest absorption peak around 588nm, which can be observed by naked eyes directly without any equipment. </p> | ||
| + | <p>We designed 2 pairs of primers for the genes of honeybee silk protein and blue chromoprotein by molecular biology technology. (There is a GGGGS linker in the middle of two fragments. ) Then we use infusion technology to insert two fragments after T7 promoter of pET21a(+) vector. The verification by PCR is showed in Figure 1. After then we introduced the recombinant plasmid into E.coli BL21(DE3). In 48 hours, there is blue chromoprotein expressed in bacterium solution, which can be observed by naked eyes (Figure 2). Therefore, the improvement of honeybee silk protein is practical and valid. </p> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/3/38/T--BNU-China--ach1.jpg" width="65%"> | ||
| + | <figcaption> | ||
| + | Fig.1 verification of recombinant plasmid by PCR. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/0/03/T--BNU-China--ach2.jpg" width="65%"> | ||
| + | <figcaption> | ||
| + | Fig.2 the bacterium solution of expression group and control group. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <p>The result of the experiment is showed in the following figures. Figure 3 showed the different colors of E.coli DL21 with different expression levels after centrifugation. There are largely expression group, slightly expression group and control group from left to right. The samples are investigated by SDS-PAGE after crushing the bacteria of three groups by high temperature. As it is showed in Figure 4 , after dyed by coomassie brilliant blue, there is a large number of protein around expected 56-57kD from largely expression group, a small number of protein from slightly expression group, and almost no significant expression from control group. The result of SDS-PAGE shows that the improved honeybee silk protein can be observed by naked eyes, and the different color shades of blue can reflect the expression level of the protein. </p> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/d/da/T--BNU-China--ach3.jpg" width="65%"> | ||
| + | <figcaption> | ||
| + | Fig.3 the bacteria of different groups after centrifugation. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/71/T--BNU-China--ach4.jpg" width="65%"> | ||
| + | <figcaption> | ||
| + | Fig.4 the result of SDS-PAGE. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p>By infusing chromoprotein with different colors after the honeybee silk protein, we can produce silk protein with different color, which will allow us to obtain the expression time, expression level and expression efficiency of the silk protein, therefore control the production of the honeybee silk protein efficiently. Not only can we make the silk protein more aesthetic, but also we can apply the silk protein into more areas widely, such as health and medicine, textile industry and biomaterial. </p> | ||
| + | <p>In this way we creat a new part: Honeybee-blue with amilCP protein (<a href='https://parts.igem.org/Part:BBa_K2220028'>BBa_K2220028</a>) </p> | ||
| + | </html> | ||
| + | <h1>Austin_UTexas 2017 Experience</h1> | ||
| + | <br> | ||
| + | <p>Due to the observed instability of the original blue chromoprotein sequence, cells expressing the blue chromoprotein have difficulty retaining the blue phenotype, as the blue chromoprotein is evolutionarily unstable due to the metabolic load imposed by its expression. In order to combat the metabolic burden and subsequent mutational inactivation of the blue chromoprotein, a technique called codon optimization was used to improve the function of this part as a reliable and stable color reporter. Codon optimization involves exchanging certain codons for ones that have known to be more translationally efficient in a certain species while retaining the same sequence of amino acids. Making the translation process more efficient, in turn, lowers the metabolic burden in the cells that the blue chromoprotein gene is present in.</p> | ||
| + | <p>To create the codon-optimized blue chromoprotein BioBrick part, a digestion was performed on the BBa_K608002 vector (containing a strong promoter and RBS) and the codon-optimized blue chromoprotein, which was ordered as a gBlock from the Integrated DNA Technologies (IDT) website. Upon digesting these parts and conducting gel extraction, a ligation reaction was used to join the codon-optimized blue chromoprotein with the BBa_K608002 vector. Following the purification of the ligation, a transformation was done via electroporation which yielded one phenotypically blue colony, shown below in <b>Figure 1.</b> This colony was then inoculated into liquid media to make an overnight culture, shown in <b>Figure 2</b>, which maintained a blue phenotype.</p> | ||
| + | <html> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/thumb/0/0f/Codon_op_blue_cp_transformation.png/247px-Codon_op_blue_cp_transformation.png"> | ||
| + | <figcaption> | ||
| + | <b>Figure 1.</b> The image above shows the transformation plate of the electroporation of the codon-optimized blue chromoprotein. The blue colony shown was used to make overnight cultures to miniprep and sequence. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/thumb/6/6f/Tubes_blue_cp.png/283px-Tubes_blue_cp.png"> | ||
| + | <figcaption> | ||
| + | <b>Figure 2.</b> On the left is the overnight culture of the phenotypically blue colony from the codon-optimized blue chromoprotein transformation. The right culture tube contains a culture of untransformed <i>E. coli</i>. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p>The codon optimized blue chromoprotein sequence with the attached RBS and promoter ((<a href="https://parts.igem.org/Part:BBa_K2253002">BBa_K2253002</a></b>)) has been sequence verified. However, there is one point mutation (5’ TTT 3’ to 5’ TTC 3’) in the nucleotide sequence, shown in <b>Figure 3.</b> Fortunately, this mutation is silent as it still retains the amino acid phenylalanine and thus the overall codon-optimized sequence of the blue chromoprotein. Another improvement of this part is that the blue chromoprotein BioBrick part now has the promoter and RBS sequence attached to it, making it more convenient for synthetic biologists to use as an additional cloning step is omitted. </p> | ||
| + | <figure class="text-center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/thumb/d/d2/Benchlingbluecp.png/800px-Benchlingbluecp.png"> | ||
| + | <figcaption> | ||
| + | <b> Figure 3.</b> Results from Benchling show the silent mutation (from 5’ TTT 3’ to 5’ TTC 3’) within the codon-optimized sequence. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | |||
| + | <h1>Uppsala 2018 Experience</h1> | ||
| + | |||
| + | These results were obtained with incorporated plasmid containing the Biobrick part [[part:BBa_K2669003]] in the IDT supplied backbone pUCIDT (Amp). | ||
| + | |||
| + | |||
| + | The original native AmilCP sequence, Part:BBa_K592009 was codon optimized for E.coli K12 with a codon optimization tool provided by Integrated DNA Technologies (IDT). In order to conduct a stability assay through growth in liquid culture, the codon optimized basic part and the original native AmilCP part was attached to a constitutive promotor, RBS and a double terminator separately. The two Biobrick parts were ordered as customized genes from IDT inserted in pUCIDT (Amp). | ||
| + | |||
| + | Both vectors were transformed into DH5-aplha E.coli cells through single tube transformation and yielded phenotypically blue colonies for cells containing plasmid with original part and plasmid containing codon optimized part (Figure 1 and 2). | ||
| + | |||
| + | === <p>Transformation results</p> === | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/7/79/T--Uppsala--T--Uppsala--Transformation_IDT_IDT-Colonies.jpeg" width="100%" height="100%"> | ||
| + | <p><b>Figure 1:</b> Transformation plate of colonies with the incorporated plasmid containing the IDT optimised original native AmilCP (BBa_K592009) sequence in the IDT supplied backbone. Colonies retrieved for the stability assay are circled.</p> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2018/2/29/T--Uppsala--Transformation_Original_IDTbb-Colonies.jpeg" width="100%" height="100%"> | ||
| + | <p><b>Figure 2:</b> Transformation plate of colonies with the incorporated plasmid containing the original native AmilCP (BBa_K592009) sequence in the IDT supplied backbone. Colonies retrieved for the assay are circled.</p> | ||
| + | </html> | ||
| + | |||
| + | === <p>Stability assay</p> === | ||
| + | <p>Stability assay through growth in liquid culture was performed with both inserts in the IDT supplied backbone. 1 mL of LB with ampicillin (25 ug/ml) was inoculated with one single colony in eppendorf tubes in 10 replicates for each part. In order to allow 10 generations of growth, a 1:1000 dilution was made of the overnight culture and incubated for 24 h. The colour comparison was done through visualization of centrifuged cultures and by analyzing the color intensity we could deduce the stability of the chromoprotein-encoding plasmid. </p> | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/e/e7/T--Uppsala--10_Gen_IDTbb_ORI_IDT.jpeg" width="100%" height="100%"> | ||
| + | <p><b>Figure 3:</b> The result after 10 generations. The upper row represents 10 different culture pellets of the cells with the plasmid containing the IDT optimized sequence of AmilCP. The lower row represents the ones with the original native AmilCP (BBa_K592009) sequence incorporated in the plasmid. </p> | ||
| + | </html> | ||
| + | |||
| + | === <p>Interpretation</p> === | ||
| + | <p> Already after 10 generations it was clearly visible that the color intensity was better distributed in the cells containing the plasmid with the IDT optimized AmilCP sequence, since all of the culture pellets had a large color intensity while the cells with the plasmid containing the original AmilCP sequence had a larger variation in color intensity. This indicates that the stability of the cells containing the plasmid with the IDT optimized AmilCP sequence is greater than those with the original AmilCP sequence. | ||
| + | |||
| + | <html> | ||
| + | <p><b>Table 1:</b> Interpretation of color expression of the cells containing the original AmilCP sequence or the IDT optimized AmilCP sequence after 10 generations of growth. ++ represents strong color, + represents weak color and - represents no color. </p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | {| class="wikitable" style="margin-left: auto; margin-right: auto; border: none; | ||
| + | |- | ||
| + | ! style="background: #98CC9A;" | Generation | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 1 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 2 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 3 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 4 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 5 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 6 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 7 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 8 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 9 | ||
| + | ! style="background: #98CC9A;" | Original AmilCP Colony 10 | ||
| + | |- | ||
| + | ! style="background: #98CC9A;" | 10 | ||
| + | | ++ | ||
| + | | + | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | + | ||
| + | | + | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | + | ||
| + | |- | ||
| + | ! style="background: #98CC9A;" | Generation | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 1 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 2 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 3 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 4 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 5 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 6 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 7 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 8 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 9 | ||
| + | ! style="background: #98CC9A;" | IDT Optimized AmilCP Colony 10 | ||
| + | |- | ||
| + | ! style="background: #98CC9A;" | 10 | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | | ++ | ||
| + | |} | ||
Latest revision as of 02:47, 18 October 2018
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K592009
User review of SHSBNU_China
Figure 1 TtrS/R system and ThsS/R system ThsS (BBa_K2507000) and ThsR (BBa_K2507001), both codon-optimized for E. coli, are two basic parts which belong to the two-component system from the marine bacterium Shewanella halifaxensis. By linking thsR with chromoprotein genes (BBa_K2507009, BBa_K2507010, BBa_K2507011) or the violacein producing operon vioABDE (BBa_K2507012), this system can respond to thiosulfate by producing a signal visible to the naked eye, such as chromoproteins (spisPink-pink chromoprotein, gfasPurple-purple chromoprotein, amilCP-blue chromoprotein) or a dark-green small-molecule pigment (protoviolaceinic acid). E. coli-codon-optimized TtrS(BBa_K2507002) and TtrR (BBa_K2507003) are two basic parts which are derived from the two-component system of the marine bacterium Shewanella baltica.
By linking ttrR with chromoprotein genes (BBa_K2507009, BBa_K2507010, BBa_K2507011) or the violacein producing operon vioABDE (BBa_K2507012), this system can respond to thiosulfate by producing a signal visible to the naked eye such as chromoproteins (spisPink-pink chromoprotein, gfasPurple-purple chromoprotein, amilCP-blue chromoprotein) or a dark-green small-molecule pigment (protoviolaceinic acid).
Figure 2. Characterization of the ThsS/R and TtrS/R system by observing the chromprotein expression levels. We added 1mM, 0.1mM, 0.01mM and 0 Na2S2O3 to ThsS/R system and added 2.5mM, 1mM, 0.1mM and 0 Na2S4O6·2H2O to ThsS/R system. The results demonstrate there is an obvious response in the ThsS/R system. In a. gfasPurple system, the response curve is obvious, and b. spisPink & c. amilCP with rather heavy leaky expression without inducer. d.e.f. However, in TtrS/R system there is no clear result.
Figure 3. Characterization of the ThsS/R and TtrS/R system by observing the protoviocaceinic acid( dark-green pigment). We added 1mM, 0.1mM, 0.01mM and 0 Na2S2O3 to ThsS/R system and added 2.5mM, 1mM, 0.1mM and 0 Na2S4O6·2H2O to ThsS/R system. The results demonstrate there is an obvious response in the ThsS/R system with rather heavy leaky expression without inducer. And the Ths/R+protoviolaceinic acid works rather well!
User Reviews
UNIQf20349fdc09deef2-partinfo-00000000-QINU
UNIQf20349fdc09deef2-partinfo-00000001-QINU
UNIQf20349fdc09deef2-partinfo-00000002-QINU
|
•••••
iGEM Sydney Australia |
Figure 1. Purple, pink and green mutants compared to the wild type.
|
|
•••••
iGEM Uppsala 2013 |
This part works well as a reporter. As expected, different levels of expression of amilCP in E. coli MG1655 generates different intensities in color. amilCP expressed from the high copy plasmid pSB1C3 by weak promoter J23116 (top part of plates) and strong promoter J23110 (bottom part of plates) at 37 °C (left plate) and 30 °C (right plate) respectively. After incubation at 30 °C a clear difference in color intensity can be seen between the expression from the two different promoters, while after incubation at 37 °C the difference in intensity is much less pronounced. This indicates that expression from the promoters are temperature dependent, and it shows that the quantifiable range of the chromoproteins is somewhat limited. Plates were incubated for 23 h at 37 °C or 30 °C followed by 20 h at 22 °C. |
UNIQf20349fdc09deef2-partinfo-00000005-QINU
|
•••••
Paris_Saclay 2013 |
Our team improved the biobrick BBa_K592009 by assembly of biobricks B0034, BBa_K592009 and B0015 to construct a composite part including a ribosome binding site, full length of amilCP gene and double terminator. We used it as one of our reporter gene in Escherichia coli to controlled activator or repressor activity of our promotor under aerobic or anaerobic conditions (Pndh*, PnirB, PnarG, PnarK, PbphR1, PbphR2). Construction : Pndh* (repressor) + BBa_K1155003 :
Tubes 1 and 2 : cellular pallet of cultures grown in aerobic conditions Tubes 3 and 4 : cellular pallet of cultures grown in anaerobic conditions |
UNIQf20349fdc09deef2-partinfo-00000007-QINU
UNIQf20349fdc09deef2-partinfo-00000008-QINU
UNIQf20349fdc09deef2-partinfo-00000009-QINU
|
•••••
iGEM Michigan 2013 |
The 2013 Michigan igem team successfully used this part to produce amilCP. Fig 2. - An inducible fim transcriptor system changes states and produces protein output NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON. |
|
•••••
Immudzen |
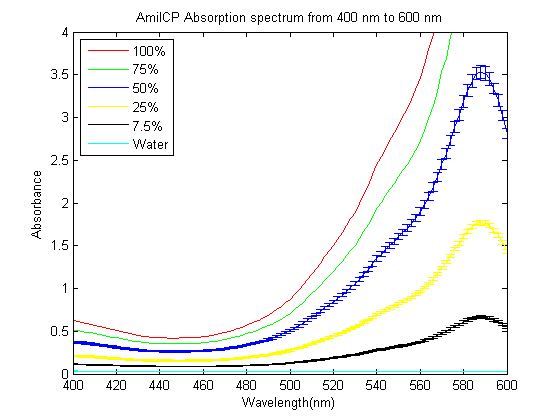
As part of the 2013 CU Boulder project we worked on separating RFP from AmilCP and we measured the spectrum of AmilCP from 400 nm to 600 nm. We also found that when running on an agarose gel RFP will run down on the gel while AmilCP runs up on the gel. |
|
•••••
iGEM 2013 Hong_Kong_HKUST Trevor Y.H. HO |
Error creating thumbnail: Invalid thumbnail parameters This part was employed in testing our Gibson Assembly protocol. In the experiment design, the mRFP coding DNA sequence in J04450 was replaced with that of amilCP. The resulting plate from transformation of the post-reaction mixture is shown. The color of the blue chromoprotein was clearly distinguished from that of mRFP, which aided in my evaluation of the performance of Gibson Assembly. |
|
BBa_K592009 iGEM Groningen 2012 |
Our team has managed to couple this biobrick part with our promoters: alsT, fnr, and sboA. The cloning was done in BBa_K818000 (plasmid backbone for B. subtilis, engineered by team Groningen 2012), to allow color expression in B. subtilis. We utilized a strong RBS BBa_B0034 for pigment expression in E. coli and B. subtilis. The purple/blue colour was strongly visible in E.coli without any induction, while the expression in B. subtilis was more subtle (B. subtilis colony looks slightly blue on the plate agar). After induction of the promoter before AmilCP, B. subtilis also turned clearly purple/blue. Please have a look at the page of BBa_K818400 for more information. |
|
No review score entered. User:agynna |
iGEM Team Uppsala University 2012 We have observed that expression of amilCP confers a noticeable fitness cost in E coli. This is surprising, given that such behaviour is not seen in other homologous chromoproteins and fluorescent proteins. We thus recommend using the new aeBlue (BBa_K864401) chromoprotein for blue color expression, which also has a more clear blue color. !!!!!!! |
|
BBa_K592009 iGEM DTU-Denmark 2017 |
The DTU-Denmark 2017 team has added a 6 x his-tag to the C-terminal end of the amilCP protein, to ensure easy purification of amilCP using affinity chromatography. The his-tagged amilCP was submitted to the parts registry as BBa_K2355002. The recombinant amilCP with his-tag was expressed in E.coli, and the and the His-tagged amilCP was purified using a Ni-NTA spin colomn. Compared to a control with expressed amilCP from BBa_K592009, the his-tagged amilCP had a 2-4 fold increase in amilCP concentration following the his-tag purification. Take a look at the entry of BBa_K2355002 for more information. |
BNU-China 2017 Experience
Team UCLA in 2015 iGEM used honeybee silk protein 3 (part number: BBa_K1763000) in their project, which can serve the purpose of being the substitution of spider thread and natural silk. Although it is not as strong as other kind of silk protein, the honeybee silk protein has some advantages compared to spider thread and natural silk: the gene of honeybee silk protein is smaller, with no duplicated sequences, and easier to be synthesized and modified by molecular biology technology.
However, the silk protein produced from UCLA has no color, thus with less aesthetic characteristic and uneasy to observe directly. In order to observe the produced honeybee silk protein more directly, we add a blue chromoprotein, amilCP (part number: BBa_K592009) , to the end of the silk protein. AmilCP is a blue chromoprotein comes from coral, which has a strong blue after expressing. The color has the biggest absorption peak around 588nm, which can be observed by naked eyes directly without any equipment.
We designed 2 pairs of primers for the genes of honeybee silk protein and blue chromoprotein by molecular biology technology. (There is a GGGGS linker in the middle of two fragments. ) Then we use infusion technology to insert two fragments after T7 promoter of pET21a(+) vector. The verification by PCR is showed in Figure 1. After then we introduced the recombinant plasmid into E.coli BL21(DE3). In 48 hours, there is blue chromoprotein expressed in bacterium solution, which can be observed by naked eyes (Figure 2). Therefore, the improvement of honeybee silk protein is practical and valid.


The result of the experiment is showed in the following figures. Figure 3 showed the different colors of E.coli DL21 with different expression levels after centrifugation. There are largely expression group, slightly expression group and control group from left to right. The samples are investigated by SDS-PAGE after crushing the bacteria of three groups by high temperature. As it is showed in Figure 4 , after dyed by coomassie brilliant blue, there is a large number of protein around expected 56-57kD from largely expression group, a small number of protein from slightly expression group, and almost no significant expression from control group. The result of SDS-PAGE shows that the improved honeybee silk protein can be observed by naked eyes, and the different color shades of blue can reflect the expression level of the protein.


By infusing chromoprotein with different colors after the honeybee silk protein, we can produce silk protein with different color, which will allow us to obtain the expression time, expression level and expression efficiency of the silk protein, therefore control the production of the honeybee silk protein efficiently. Not only can we make the silk protein more aesthetic, but also we can apply the silk protein into more areas widely, such as health and medicine, textile industry and biomaterial.
In this way we creat a new part: Honeybee-blue with amilCP protein (BBa_K2220028)
Austin_UTexas 2017 Experience
Due to the observed instability of the original blue chromoprotein sequence, cells expressing the blue chromoprotein have difficulty retaining the blue phenotype, as the blue chromoprotein is evolutionarily unstable due to the metabolic load imposed by its expression. In order to combat the metabolic burden and subsequent mutational inactivation of the blue chromoprotein, a technique called codon optimization was used to improve the function of this part as a reliable and stable color reporter. Codon optimization involves exchanging certain codons for ones that have known to be more translationally efficient in a certain species while retaining the same sequence of amino acids. Making the translation process more efficient, in turn, lowers the metabolic burden in the cells that the blue chromoprotein gene is present in.
To create the codon-optimized blue chromoprotein BioBrick part, a digestion was performed on the BBa_K608002 vector (containing a strong promoter and RBS) and the codon-optimized blue chromoprotein, which was ordered as a gBlock from the Integrated DNA Technologies (IDT) website. Upon digesting these parts and conducting gel extraction, a ligation reaction was used to join the codon-optimized blue chromoprotein with the BBa_K608002 vector. Following the purification of the ligation, a transformation was done via electroporation which yielded one phenotypically blue colony, shown below in Figure 1. This colony was then inoculated into liquid media to make an overnight culture, shown in Figure 2, which maintained a blue phenotype.


The codon optimized blue chromoprotein sequence with the attached RBS and promoter ((BBa_K2253002)) has been sequence verified. However, there is one point mutation (5’ TTT 3’ to 5’ TTC 3’) in the nucleotide sequence, shown in Figure 3. Fortunately, this mutation is silent as it still retains the amino acid phenylalanine and thus the overall codon-optimized sequence of the blue chromoprotein. Another improvement of this part is that the blue chromoprotein BioBrick part now has the promoter and RBS sequence attached to it, making it more convenient for synthetic biologists to use as an additional cloning step is omitted.

Uppsala 2018 Experience
These results were obtained with incorporated plasmid containing the Biobrick part part:BBa_K2669003 in the IDT supplied backbone pUCIDT (Amp).
The original native AmilCP sequence, Part:BBa_K592009 was codon optimized for E.coli K12 with a codon optimization tool provided by Integrated DNA Technologies (IDT). In order to conduct a stability assay through growth in liquid culture, the codon optimized basic part and the original native AmilCP part was attached to a constitutive promotor, RBS and a double terminator separately. The two Biobrick parts were ordered as customized genes from IDT inserted in pUCIDT (Amp).
Both vectors were transformed into DH5-aplha E.coli cells through single tube transformation and yielded phenotypically blue colonies for cells containing plasmid with original part and plasmid containing codon optimized part (Figure 1 and 2).
Transformation results

Figure 1: Transformation plate of colonies with the incorporated plasmid containing the IDT optimised original native AmilCP (BBa_K592009) sequence in the IDT supplied backbone. Colonies retrieved for the stability assay are circled.

Figure 2: Transformation plate of colonies with the incorporated plasmid containing the original native AmilCP (BBa_K592009) sequence in the IDT supplied backbone. Colonies retrieved for the assay are circled.
Stability assay
Stability assay through growth in liquid culture was performed with both inserts in the IDT supplied backbone. 1 mL of LB with ampicillin (25 ug/ml) was inoculated with one single colony in eppendorf tubes in 10 replicates for each part. In order to allow 10 generations of growth, a 1:1000 dilution was made of the overnight culture and incubated for 24 h. The colour comparison was done through visualization of centrifuged cultures and by analyzing the color intensity we could deduce the stability of the chromoprotein-encoding plasmid.

Figure 3: The result after 10 generations. The upper row represents 10 different culture pellets of the cells with the plasmid containing the IDT optimized sequence of AmilCP. The lower row represents the ones with the original native AmilCP (BBa_K592009) sequence incorporated in the plasmid.
Interpretation
Already after 10 generations it was clearly visible that the color intensity was better distributed in the cells containing the plasmid with the IDT optimized AmilCP sequence, since all of the culture pellets had a large color intensity while the cells with the plasmid containing the original AmilCP sequence had a larger variation in color intensity. This indicates that the stability of the cells containing the plasmid with the IDT optimized AmilCP sequence is greater than those with the original AmilCP sequence.
Table 1: Interpretation of color expression of the cells containing the original AmilCP sequence or the IDT optimized AmilCP sequence after 10 generations of growth. ++ represents strong color, + represents weak color and - represents no color.
| Generation | Original AmilCP Colony 1 | Original AmilCP Colony 2 | Original AmilCP Colony 3 | Original AmilCP Colony 4 | Original AmilCP Colony 5 | Original AmilCP Colony 6 | Original AmilCP Colony 7 | Original AmilCP Colony 8 | Original AmilCP Colony 9 | Original AmilCP Colony 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | ++ | + | ++ | ++ | + | + | ++ | ++ | ++ | + |
| Generation | IDT Optimized AmilCP Colony 1 | IDT Optimized AmilCP Colony 2 | IDT Optimized AmilCP Colony 3 | IDT Optimized AmilCP Colony 4 | IDT Optimized AmilCP Colony 5 | IDT Optimized AmilCP Colony 6 | IDT Optimized AmilCP Colony 7 | IDT Optimized AmilCP Colony 8 | IDT Optimized AmilCP Colony 9 | IDT Optimized AmilCP Colony 10 |
| 10 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |