Difference between revisions of "Part:BBa K1150020"
| (29 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1150020 short</partinfo> | <partinfo>BBa_K1150020 short</partinfo> | ||
| + | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="right" | ||
| + | ! colspan="2" style="background:#FFBF00;"|CMV:Ha-NLS-dCas9-Linker-VP16-NLS:BGH | ||
| + | |- | ||
| + | |'''Function''' | ||
| + | |gene activation | ||
| + | |- | ||
| + | |'''Use in''' | ||
| + | |Prokaryotic cells | ||
| + | |- | ||
| + | |'''RFC standard''' | ||
| + | |[https://parts.igem.org/Help:Assembly_standard_10 RFC 10], RFC 10 compatible | ||
| + | |- | ||
| + | |'''Backbone''' | ||
| + | |pSB1C3<br> | ||
| + | |- | ||
| + | |'''Submitted by''' | ||
| + | |[http://2016.igem.org/Team:NEU-China] | ||
| + | |} | ||
| − | |||
| − | The | + | The Freiburg iGEM team 2013 designed a fusion protein consisting of dCas9 and VP16 [1-6] for sequence-specific transactivation of a desired target locus [http://2013.igem.org/Team:Freiburg/Project/effector#activation (more information)]. Therefore, we used our double mutated dCas9 [https://parts.igem.org/Part:BBa_K1150000 (BBa_K1150000)] impaired in its cleavage activity and fused it to the 5’ end of the sequence coding for the transactivation domain of VP16 [https://parts.igem.org/Part:BBa_K1150001 (BBa_K1150001)]. To ensure nuclear localization of the construct a nuclear localization signal (NLS) was fused to both ends of dCas9-VP16. For detection of expression the fusion protein was tagged with a HA-epitope coding sequence [https://parts.igem.org/Part:BBa_K1150016 (BBaa_K1150016)] and its expression was set under control of the CMV promoter [https://parts.igem.org/Part:BBa_K747096 (BBa_K747096)] and BGH terminator [https://parts.igem.org/Part:BBa_K1150012 (BBa_K1150012)]. Figure 1 illustrates the detailed design of the whole device. |
| − | + | [[File:Overview_of_the_construct_CMV-Cas-VP16_Freigem_2013.png|700px|]] | |
| − | ''' | + | '''Figure 1: Construct design.''' dCas9 was fused via a 3 amino acid linker to VP16. The resulting fusion construct was flanked by NLS sequences and tagged by a HA epitope. The CMV promoter and BGH terminator were chosen to control gene expression. |
| − | + | ||
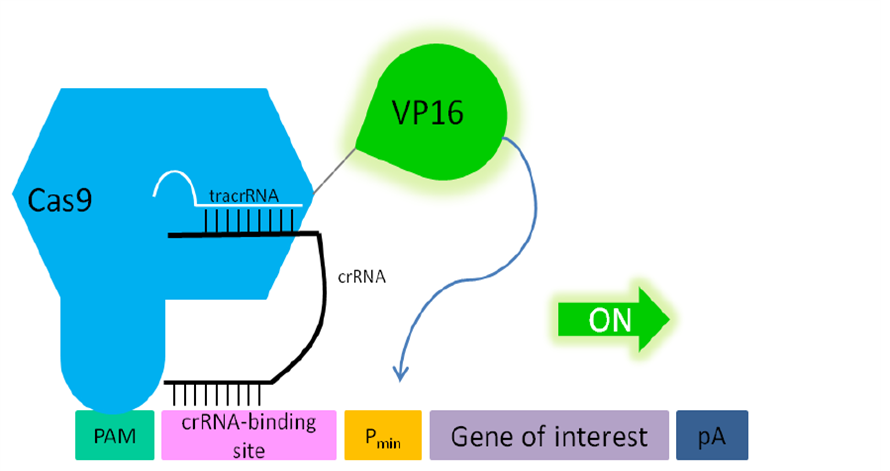
| − | + | By co-transfecting our RNA plasmid [https://parts.igem.org/Part:BBa_K1150034 (BBa_K1150034)] which includes the tracrRNA and the separately integrated, desired crRNA, the dCas9 specifically binds to the targeted DNA sequence. With the help of the transactivation domain of VP16, transcription factors are recruited and the pre-initiation complex can be built. By placing this construct upstream of a promotor region any gene of interest can be activated. | |
| − | For testing | + | |
| − | Different crRNAs were tested | + | [[File:Picture1VP16-Cas_Freigem2013.png|700px|]] |
| + | |||
| + | '''Figure 2: Principle of transactivation of mammalian gene expression by the fusion protein dCas9-VP19 (BBa_K1150020).''' The double mutated dCas9 (D10A; H840A) fused to the herpes simplex virus (HSV) derived VP16 activation domain can serve as a crRNA-guided DNA-binding and transactivating protein. If a PAM sequence is present at the 3’ end of the crRNA binding site almost any DNA sequence can be targeted. Abbr.: Pmin: minimal promoter, containing minimal requirements for binding of transcription factors; Cas9: CRISPR associated protein 9; crRNA: CRISPR RNA; tracrRNA: trans-activating RNA; VP16: Virus protein 16, herpes simplex virus (HSV)-derived transcriptional activator protein; PAM: protospacer adjacent motif. | ||
| + | |||
| + | |||
| + | |||
| + | ===Experimental approach=== | ||
| + | |||
| + | For testing this device we used HEK-293T cells, which were seeded at a densitiy of 65,000 cells/well in 24-well plates. After 24 hours RNA plasmids with crRNAs against the indicated loci and the referring reporter plasmids containing a gene coding for the secreted alkaline phosphatase (SEAP) under the control of a CMV minimal promoter were co-transfected to this device. 48 hours post transfection the activity of SEAP in the cell culture medium was measured. Additionally, the dCas9-VP16 expression was assessed by Western blot analysis of cell lysates. Different crRNAs were tested and compared for their activation properties of the referring reporter plasmid. | ||
==Proof of function== | ==Proof of function== | ||
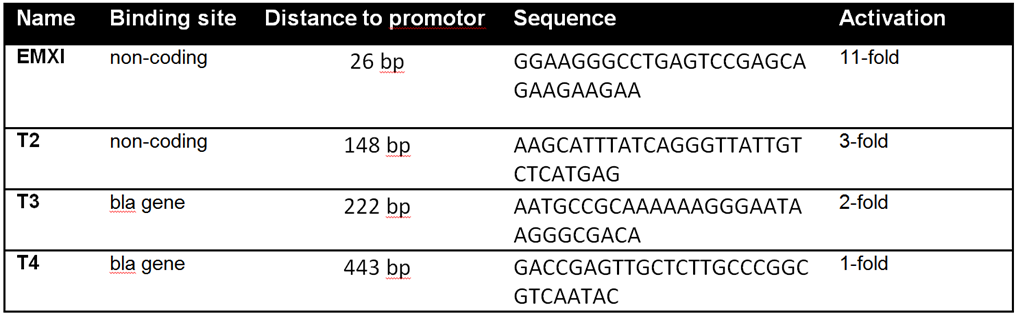
| − | With BBa_K1150020 different target loci | + | With BBa_K1150020 different target loci have been tested by the usage of a SEAP reporter plasmid with a minimal CMV promotor. The target sites can be determined by directing the crRNA consisting of 30 bp length against the desired sequence of interest. T2 [https://parts.igem.org/Part:BBa_K1150040 (BBa_K1150040)] and EMXI [https://parts.igem.org/Part:BBa_K1150035 (BBa_K1150035)] with target sites at different distances to the promotor regions proved successfully as potential activation sites (see Table 1 and Figure 3). |
| − | + | ||
| − | Table | + | '''Table 1:''' Overview of the tested crRNAs with different binding sites on the SEAP plasmid. |
| + | [[File:CrRNAs_for_activation_Cas-VP16_Freigem2013.png|700px|]] | ||
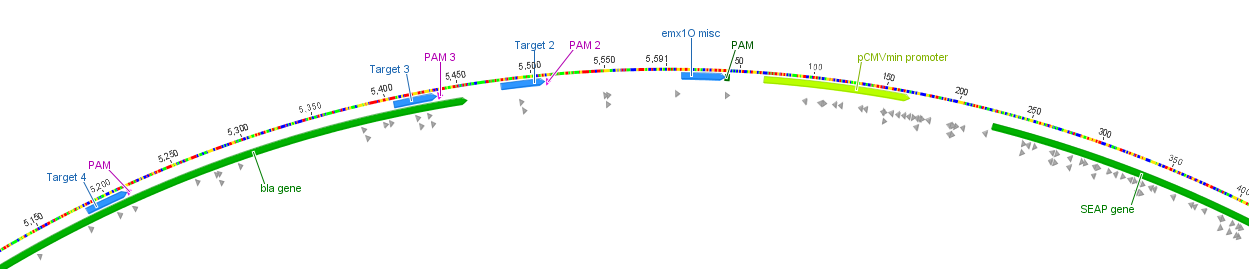
| + | |||
| + | [[File:Activation_targets_on_SEAP_plasmid_Freigem_2013.png|700px|]] | ||
| + | |||
| + | '''Figure 3:''' Position of the target loci on the SEAP plasmid. | ||
| + | |||
| + | |||
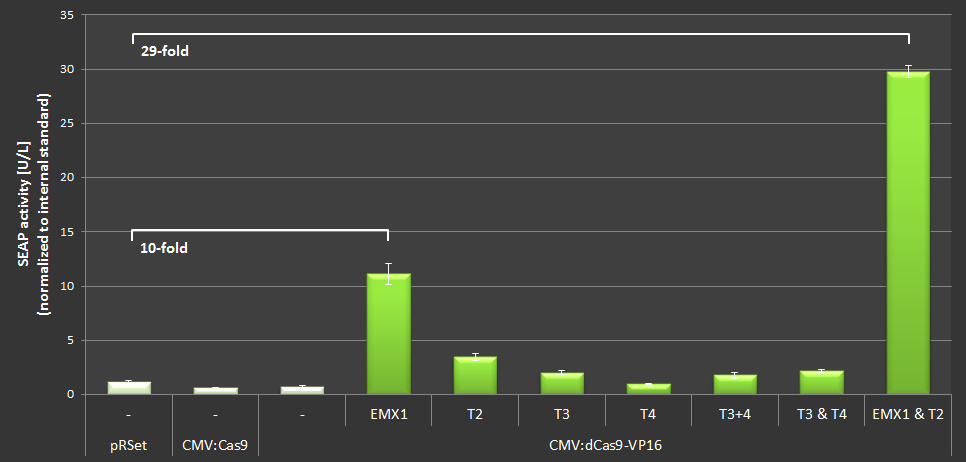
| + | To quantify the activation properties of dCas9-VP16 (BBa_K1150020) the amount of SEAP expression was measured and compared to the basis-SEAP-level expressed by the CMVmin promotor. Each sample was measured in biological triplicates. The results are listed in Figure 4. By simultaniously using the EMXI and T2 loci an approximately 28-fold increase in SEAP production could be determined. | ||
| + | |||
| + | [[File:CMV-Cas-VP16_Results_Freigem2013.png|900px|]] | ||
| + | |||
| + | '''Figure 4:''' Results of the SEAP-activation with dCas-VP16 under the CMV promotor using different crRNAs. | ||
| + | |||
| + | =Activation of a fluorescence reporter= | ||
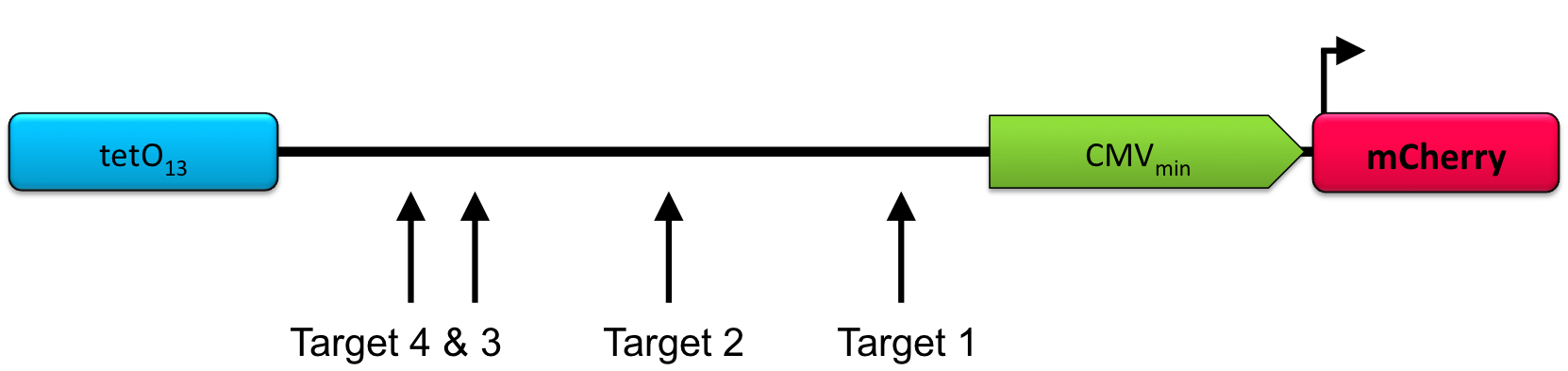
| + | To prove broad functionality of this device we activated a CMVmin:mCherry with it. Again we were using different RNA plasmids that adress positions in front of the promoter. | ||
| + | |||
| + | [[File:Mcherry_Freiburg_2013.PNG|700px|]] | ||
| + | |||
| + | '''Figure 5:'''Position of the target sites in front of the mCherry reporter construct. | ||
| + | |||
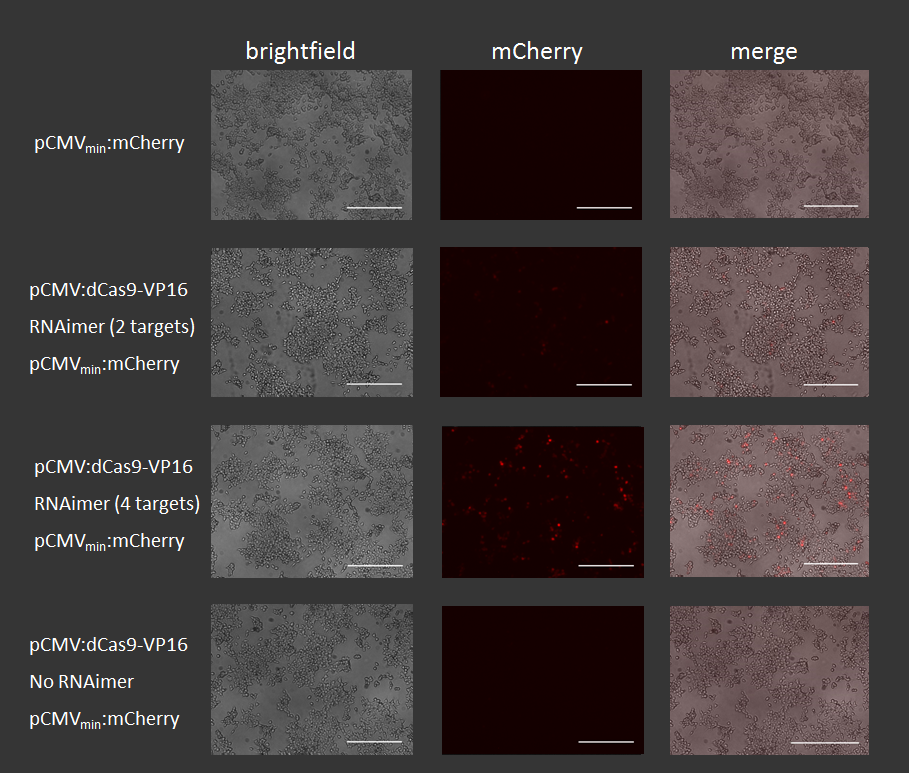
| + | HEK-293T cells were transfected with the reporter construct, dCas9-VP16 and RNAimer plasmids coding for crRNAs that adress different positions in front of the minimal promoter. 48 hours post transfection fluorescence signal was measured using an AxioVert200 microscope with the same exposure time (1000ms). Scale bar = 200µm. | ||
| + | |||
| + | [[File:Results_micorscoe_Freiburg_PS_2013.PNG|700px|]] | ||
| + | |||
| + | '''Figure 6:''' Results of mCherry activation via dCas9-VP16 using different crRNAs. Scale bar = 200µm. | ||
==Proof of expression== | ==Proof of expression== | ||
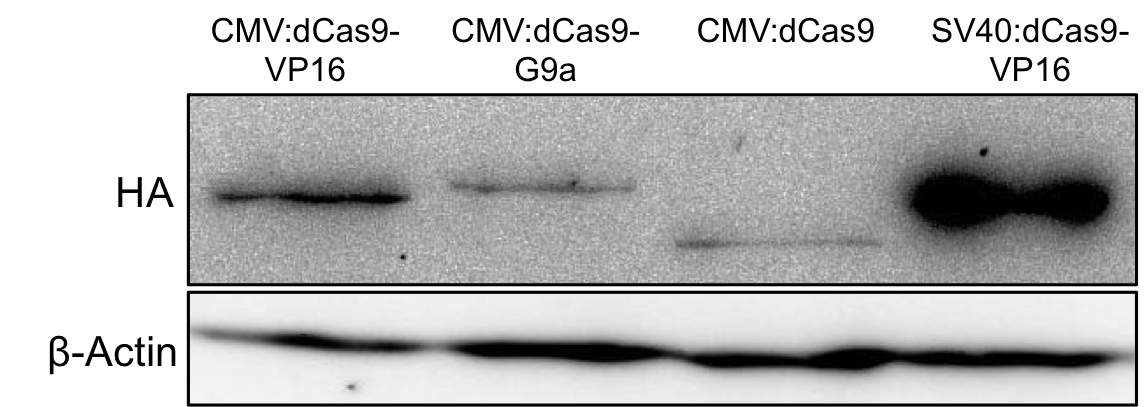
| − | With the help of the HA-tag we performed Western blots to verify the expression of BBa_K1150020 and to estimate its expression rate. | + | With the help of the HA-tag we performed Western blots to verify the expression of BBa_K1150020 and to estimate its expression rate (Fig. 7). |
| + | [[File:UniBAssblotfertig2.1-team_Freiburgforregistry.PNG|800px|]] | ||
| − | + | '''Figure 7:''' HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone. | |
| − | <span class='h3bb'>Sequence and Features</span> | + | |
| + | |||
| + | <span class='h3bb'> | ||
| + | ==Sequence and Features== | ||
| + | </span> | ||
<partinfo>BBa_K1150020 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1150020 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | == References == | ||
| + | <small> | ||
| + | |||
| + | [1] WEIR, J. (2001). Regulation of herpes simplex virus gene expression. ''Gene'' 271: 117-130. | ||
| + | |||
| + | [2] GREAVES, R.; O’HARE, P. (1989). Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65.'' J Virol'' 63: 1641-1650. | ||
| + | |||
| + | [3] TRIEZENBERG, S. J.; KINGSBURY, R. C.; MCKNIGHT, S. L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. ''Genes Dev'' 2: 718-729. | ||
| + | |||
| + | [4] HIRAI, H.; TANI, T; KIKYO, N. (2010). Structure and functions of powerful transactivators: VP16, MyoD and FoxA. ''Int. J. Dev. Biol.'' 54: 1589-1596. | ||
| + | |||
| + | [5] WALKER, R.; GREAVES, R.; O’HARE, P. (1993). Transcriptional activation by the acidic domain and involves additional determinants distinctof Vmw65 requires the integrity of the domain from those necessary for TFIIB binding. ''Molecular and Cellular Biology'' 13: 5233-5244. | ||
| + | |||
| + | [6] THOMPSON, R.; PRESTON, C.; SAWTELLl, N. (2009). De Novo Synthesis of VP16 Coordinates the Exit from HSV Latency In Vivo. ''PLoS Pathog'' 5: e1000352. | ||
| + | </small> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 05:54, 12 October 2016
uniCAS Activator (CMV promoter)
| CMV:Ha-NLS-dCas9-Linker-VP16-NLS:BGH | |
|---|---|
| Function | gene activation |
| Use in | Prokaryotic cells |
| RFC standard | RFC 10, RFC 10 compatible |
| Backbone | pSB1C3 |
| Submitted by | [http://2016.igem.org/Team:NEU-China] |
The Freiburg iGEM team 2013 designed a fusion protein consisting of dCas9 and VP16 [1-6] for sequence-specific transactivation of a desired target locus [http://2013.igem.org/Team:Freiburg/Project/effector#activation (more information)]. Therefore, we used our double mutated dCas9 (BBa_K1150000) impaired in its cleavage activity and fused it to the 5’ end of the sequence coding for the transactivation domain of VP16 (BBa_K1150001). To ensure nuclear localization of the construct a nuclear localization signal (NLS) was fused to both ends of dCas9-VP16. For detection of expression the fusion protein was tagged with a HA-epitope coding sequence (BBaa_K1150016) and its expression was set under control of the CMV promoter (BBa_K747096) and BGH terminator (BBa_K1150012). Figure 1 illustrates the detailed design of the whole device.
Figure 1: Construct design. dCas9 was fused via a 3 amino acid linker to VP16. The resulting fusion construct was flanked by NLS sequences and tagged by a HA epitope. The CMV promoter and BGH terminator were chosen to control gene expression.
By co-transfecting our RNA plasmid (BBa_K1150034) which includes the tracrRNA and the separately integrated, desired crRNA, the dCas9 specifically binds to the targeted DNA sequence. With the help of the transactivation domain of VP16, transcription factors are recruited and the pre-initiation complex can be built. By placing this construct upstream of a promotor region any gene of interest can be activated.
Figure 2: Principle of transactivation of mammalian gene expression by the fusion protein dCas9-VP19 (BBa_K1150020). The double mutated dCas9 (D10A; H840A) fused to the herpes simplex virus (HSV) derived VP16 activation domain can serve as a crRNA-guided DNA-binding and transactivating protein. If a PAM sequence is present at the 3’ end of the crRNA binding site almost any DNA sequence can be targeted. Abbr.: Pmin: minimal promoter, containing minimal requirements for binding of transcription factors; Cas9: CRISPR associated protein 9; crRNA: CRISPR RNA; tracrRNA: trans-activating RNA; VP16: Virus protein 16, herpes simplex virus (HSV)-derived transcriptional activator protein; PAM: protospacer adjacent motif.
Experimental approach
For testing this device we used HEK-293T cells, which were seeded at a densitiy of 65,000 cells/well in 24-well plates. After 24 hours RNA plasmids with crRNAs against the indicated loci and the referring reporter plasmids containing a gene coding for the secreted alkaline phosphatase (SEAP) under the control of a CMV minimal promoter were co-transfected to this device. 48 hours post transfection the activity of SEAP in the cell culture medium was measured. Additionally, the dCas9-VP16 expression was assessed by Western blot analysis of cell lysates. Different crRNAs were tested and compared for their activation properties of the referring reporter plasmid.
Proof of function
With BBa_K1150020 different target loci have been tested by the usage of a SEAP reporter plasmid with a minimal CMV promotor. The target sites can be determined by directing the crRNA consisting of 30 bp length against the desired sequence of interest. T2 (BBa_K1150040) and EMXI (BBa_K1150035) with target sites at different distances to the promotor regions proved successfully as potential activation sites (see Table 1 and Figure 3).
Table 1: Overview of the tested crRNAs with different binding sites on the SEAP plasmid.
Figure 3: Position of the target loci on the SEAP plasmid.
To quantify the activation properties of dCas9-VP16 (BBa_K1150020) the amount of SEAP expression was measured and compared to the basis-SEAP-level expressed by the CMVmin promotor. Each sample was measured in biological triplicates. The results are listed in Figure 4. By simultaniously using the EMXI and T2 loci an approximately 28-fold increase in SEAP production could be determined.
Figure 4: Results of the SEAP-activation with dCas-VP16 under the CMV promotor using different crRNAs.
Activation of a fluorescence reporter
To prove broad functionality of this device we activated a CMVmin:mCherry with it. Again we were using different RNA plasmids that adress positions in front of the promoter.
Figure 5:Position of the target sites in front of the mCherry reporter construct.
HEK-293T cells were transfected with the reporter construct, dCas9-VP16 and RNAimer plasmids coding for crRNAs that adress different positions in front of the minimal promoter. 48 hours post transfection fluorescence signal was measured using an AxioVert200 microscope with the same exposure time (1000ms). Scale bar = 200µm.
Figure 6: Results of mCherry activation via dCas9-VP16 using different crRNAs. Scale bar = 200µm.
Proof of expression
With the help of the HA-tag we performed Western blots to verify the expression of BBa_K1150020 and to estimate its expression rate (Fig. 7).
Figure 7: HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 576
Illegal BglII site found at 900 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1] WEIR, J. (2001). Regulation of herpes simplex virus gene expression. Gene 271: 117-130.
[2] GREAVES, R.; O’HARE, P. (1989). Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J Virol 63: 1641-1650.
[3] TRIEZENBERG, S. J.; KINGSBURY, R. C.; MCKNIGHT, S. L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev 2: 718-729.
[4] HIRAI, H.; TANI, T; KIKYO, N. (2010). Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 54: 1589-1596.
[5] WALKER, R.; GREAVES, R.; O’HARE, P. (1993). Transcriptional activation by the acidic domain and involves additional determinants distinctof Vmw65 requires the integrity of the domain from those necessary for TFIIB binding. Molecular and Cellular Biology 13: 5233-5244.
[6] THOMPSON, R.; PRESTON, C.; SAWTELLl, N. (2009). De Novo Synthesis of VP16 Coordinates the Exit from HSV Latency In Vivo. PLoS Pathog 5: e1000352.