Difference between revisions of "Part:BBa B0015"
| (9 intermediate revisions by 7 users not shown) | |||
| Line 8: | Line 8: | ||
<partinfo>BBa_B0015 short</partinfo> | <partinfo>BBa_B0015 short</partinfo> | ||
* Double terminator consisting of BBa_B0010 and BBa_B0012. | * Double terminator consisting of BBa_B0010 and BBa_B0012. | ||
| − | * This is the most commonly used terminator. It seems to be reliable. | + | * This is the most commonly used terminator. It seems to be reliable.<br> Note, however, that [[Part:BBa_B0014]] is a better part for forward and reverse termination. |
<hr> | <hr> | ||
| Line 19: | Line 19: | ||
[[Image:Mfold-B0015-1.png]] | [[Image:Mfold-B0015-1.png]] | ||
| + | |||
| + | |||
| + | {| width=50% style='border: 2px solid red' | ||
| + | |- | ||
| + | |'''Using primers VR/VF2 to PCR B0010 will result in excess bands.''' | ||
| + | '''See a full description of the problem [[Problems with PCR using VR/VF2|here]].''' | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | <partinfo>BBa_B0015 SequenceAndFeatures</partinfo> | ||
<hr> | <hr> | ||
'''Measurement''' | '''Measurement''' | ||
* [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured] | * [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured] | ||
| + | |||
| + | |||
| + | |||
| + | >Internal Priming Screening Characterization of BBa_B0015: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer. | ||
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_B0015 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 5.0 ± 19.2% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | ==KEYSTONE 2022 Characterization== | ||
| + | This is one of the most commonly used terminators and is also one of the most reliable ones. This was used with several CDS such as ClSS_S533A (Part Number: BBa_K4274000) and ERG20_F96W (Part Number: BBa_K4274002). Utilization of this terminator allowed us to produce santalene from <i>E.coli</i> DH5α. | ||
| + | |||
| + | ===Santalene production in <i>E.coli</i> DH5α=== | ||
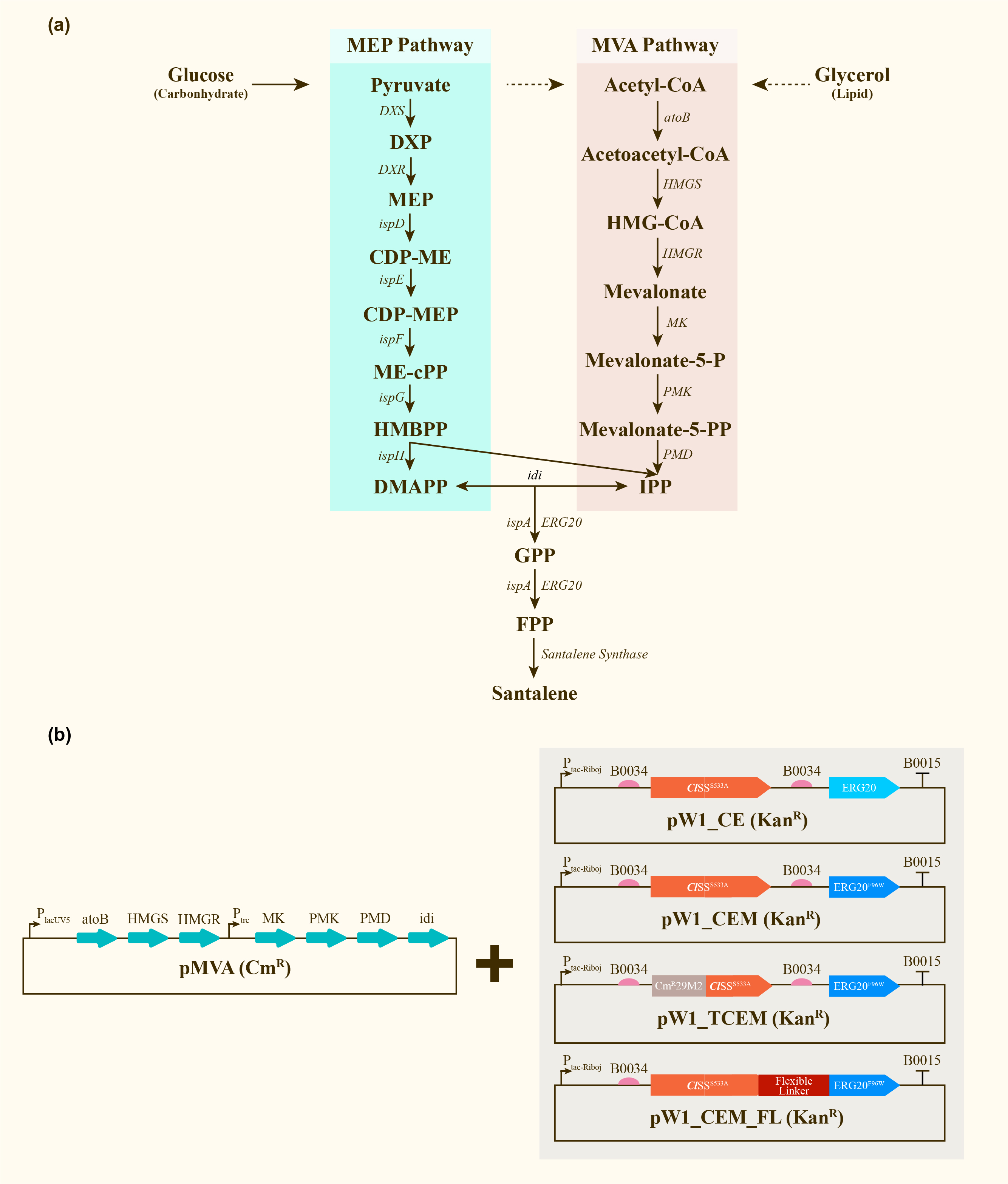
| + | After engineering, <i>E. coli</i> could utilize both MEP pathway and MVA pathway for the universal precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), then synthesize santalene with the help of FPP Synthase (FPPS) and santalene synthase (SS). Except heterologously expressed MVA pathway and ERG20 of <i>Saccharomyces cerevisiae</i> and santalene synthase of <i>Clausena lansium</i> (<i>Cl</i>SS), several modifications upon ERG20 or <i>Cl</i>SS by amino acid mutation, binding to a hydrophillic tag and the construction of fusion protein were tested for the higher yield of santalene. Therefore, with the help of the co-transformation of pMVA plasmid with various pW1 plasmids, including pW1_CE, pW1_CEM, pW1_TCEM and pW1_CEM_FL, different strains like CE, CEM, TCEM, CEM_FL were successfully constructed (Figure 1). The complete pathway we designed for producing santalene in <i>E. coli</i> is illustrated in Figure 1. | ||
| + | |||
| + | [[Image:Parts-keystone-santalene1.jpeg|thumbnail|750px|center|'''Figure 1:''' | ||
| + | Construction and expression of santalene. (a) Enzymes and some of the reaction intermediates necessary for the production of santalene through the MEP pathway and MVA pathway. (b) Schematic representing the structure of pMVA, pW1_CE, pW1_CEM, pW1_TCEM and pW1_CEM_FL transformed into <i>E.coli</i> DH5α ∆TnaA. ]] | ||
| + | |||
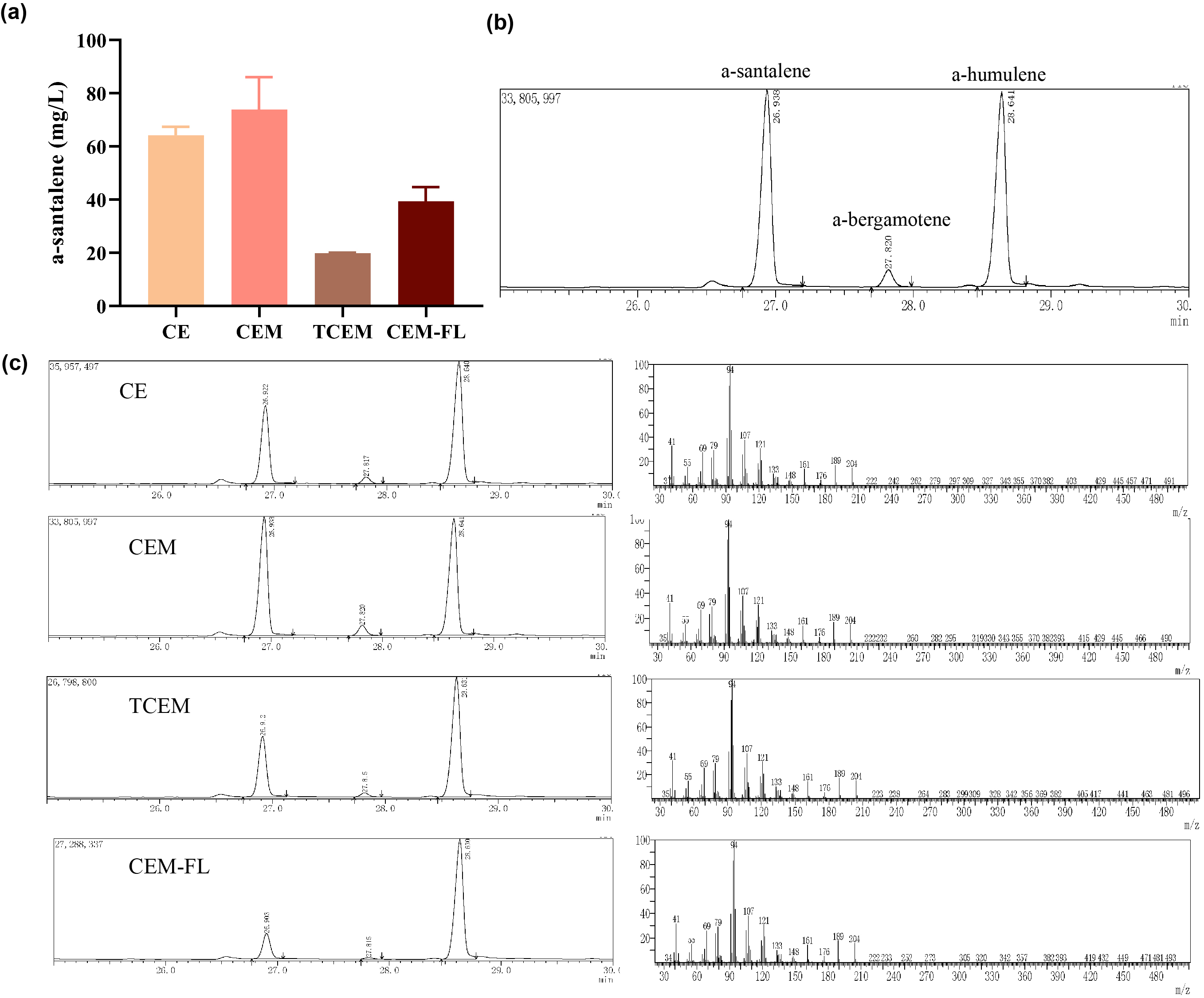
| + | Afterwards, the various engineering of E.coli DH5α ∆TnaA mentioned above were used for santalene production. After rapid centrifugation, the supernatant of dodecane was spiked with with 0.475 g/L a-humulene as an internal standard, and then injected into GC/MS for verification of α-santalene production. It turned out that all samples from four strains appeared a significant peak at the retention time of 26-27 min, and various peak area of different samples exhibited santalene production with differing levels, indicating the general success of E. coli engineering. It can be concluded that the E. coli strain CEM (with pW1_CEM plasmid) produces the maximal level of α-santalene compared to other strains (73.93 mg/L). Furthermore, our study elucidates that the mutation of 96th amino acid into tryptophan could increase the yield of α-santalene by about 20%, substantiating the prominent performance of ERG20F96W in enhancing the supply of FPP and α-santalene production in E. coli (Figure 2). | ||
| + | |||
| + | [[Image:Parts-keystone-santalene2.jpeg|thumbnail|750px|center|'''Figure 2:''' | ||
| + | Quantification analysis of α-santalene production. (a) Measurement santalene production of different strains by GC/MS results. (b) Quantification of α-santalene is analyzed with 0.475 g/L α-humulene as an internal standard. And the GC/MS results demonstrate that the peaks at the retention time of 26-27 min and 28-29 min respectively were α-santalene and α-humulene. (c) GC/MS results of samples originated from CE, CEM, TCEM and CEM-FL strains.]] | ||
| + | |||
| + | ===Contribution: New documentation from Evry_Paris-Saclay 2020=== | ||
| + | |||
| + | BBa_B0015 is a composite terminator made by joining 2 other terminators, one derived from E. coli (BBa_B0010) and the other from the T7 phage (BBa_B0012). Unlike what one can guess from the name and origin, the E. coli terminator can terminate transcription by T7 RNA polymerase, but not the T7 TE terminator. Indeed, BBa_B0010 is the E. coli rrnB T1 terminator which was shown to be an efficient terminator for the E. coli RNA polymerase, but also for the phage SP6 and T7 RNA polymerases through two different mechanism: one involving an upstream hairpin structure and the other one a downstream sequence-specific signal [1–3]. However, the T7 TE terminator (BBa_B0012) is located at the end of the T7 DNA ligase gene which is in the early region of bacteriophage T7 genome [4]. This terminator is an efficient one for the E. coli RNA polymerase, but not for the T7 RNA polymerase [5]. | ||
| + | |||
| + | ===Contribution of SCAU-China=== | ||
| + | We use this element to construct our lysis device. See more detail in '''Part:BBa_K4632024'''[https://parts.igem.org/Part:BBa_K4632024] | ||
| + | |||
| + | =====References===== | ||
| + | |||
| + | [1] Christiansen J. The 9S RNA precursor of Escherichia coli 5S RNA has three structural domains: implications for processing. Nucleic Acids Research (1988) 16: 7457–7476. | ||
| + | |||
| + | [2] Hartvig L, Christiansen J. Intrinsic termination of T7 RNA polymerase mediated by either RNA or DNA. The EMBO journal (1996) 15: 4767–4774. | ||
| + | |||
| + | [3] Kwon YS, Kang C. Bipartite modular structure of intrinsic, RNA hairpin-independent termination signal for phage RNA polymerases. The Journal of Biological Chemistry (1999) 274: 29149–29155. | ||
| + | |||
| + | [4] Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. Journal of Molecular Biology (1983) 166: 477–535. | ||
| + | |||
| + | [5] Dunn JJ, Studier FW. The transcription termination site at the end of the early region of bacteriophage T7 DNA. Nucleic Acids Research (1980) 8: 2119–2132. | ||
Latest revision as of 00:31, 11 October 2023
|
double terminator (B0010-B0012)
|
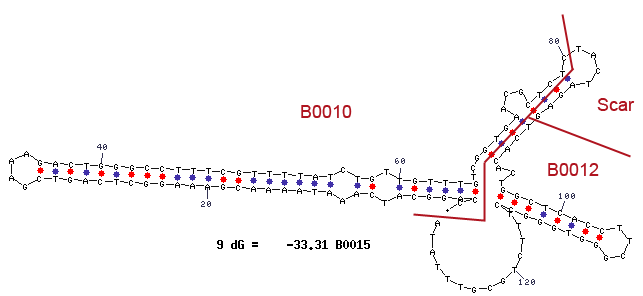
Secondary Structure
The mfold results are annotated with the location of the subparts BBa_B0010 and BBa_B0012 and the BioBrick assembly scar.
| Using primers VR/VF2 to PCR B0010 will result in excess bands.
See a full description of the problem here. |
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Measurement
- [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured]
>Internal Priming Screening Characterization of BBa_B0015: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
KEYSTONE 2022 Characterization
This is one of the most commonly used terminators and is also one of the most reliable ones. This was used with several CDS such as ClSS_S533A (Part Number: BBa_K4274000) and ERG20_F96W (Part Number: BBa_K4274002). Utilization of this terminator allowed us to produce santalene from E.coli DH5α.
Santalene production in E.coli DH5α
After engineering, E. coli could utilize both MEP pathway and MVA pathway for the universal precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), then synthesize santalene with the help of FPP Synthase (FPPS) and santalene synthase (SS). Except heterologously expressed MVA pathway and ERG20 of Saccharomyces cerevisiae and santalene synthase of Clausena lansium (ClSS), several modifications upon ERG20 or ClSS by amino acid mutation, binding to a hydrophillic tag and the construction of fusion protein were tested for the higher yield of santalene. Therefore, with the help of the co-transformation of pMVA plasmid with various pW1 plasmids, including pW1_CE, pW1_CEM, pW1_TCEM and pW1_CEM_FL, different strains like CE, CEM, TCEM, CEM_FL were successfully constructed (Figure 1). The complete pathway we designed for producing santalene in E. coli is illustrated in Figure 1.
Afterwards, the various engineering of E.coli DH5α ∆TnaA mentioned above were used for santalene production. After rapid centrifugation, the supernatant of dodecane was spiked with with 0.475 g/L a-humulene as an internal standard, and then injected into GC/MS for verification of α-santalene production. It turned out that all samples from four strains appeared a significant peak at the retention time of 26-27 min, and various peak area of different samples exhibited santalene production with differing levels, indicating the general success of E. coli engineering. It can be concluded that the E. coli strain CEM (with pW1_CEM plasmid) produces the maximal level of α-santalene compared to other strains (73.93 mg/L). Furthermore, our study elucidates that the mutation of 96th amino acid into tryptophan could increase the yield of α-santalene by about 20%, substantiating the prominent performance of ERG20F96W in enhancing the supply of FPP and α-santalene production in E. coli (Figure 2).
Contribution: New documentation from Evry_Paris-Saclay 2020
BBa_B0015 is a composite terminator made by joining 2 other terminators, one derived from E. coli (BBa_B0010) and the other from the T7 phage (BBa_B0012). Unlike what one can guess from the name and origin, the E. coli terminator can terminate transcription by T7 RNA polymerase, but not the T7 TE terminator. Indeed, BBa_B0010 is the E. coli rrnB T1 terminator which was shown to be an efficient terminator for the E. coli RNA polymerase, but also for the phage SP6 and T7 RNA polymerases through two different mechanism: one involving an upstream hairpin structure and the other one a downstream sequence-specific signal [1–3]. However, the T7 TE terminator (BBa_B0012) is located at the end of the T7 DNA ligase gene which is in the early region of bacteriophage T7 genome [4]. This terminator is an efficient one for the E. coli RNA polymerase, but not for the T7 RNA polymerase [5].
Contribution of SCAU-China
We use this element to construct our lysis device. See more detail in Part:BBa_K4632024[1]
References
[1] Christiansen J. The 9S RNA precursor of Escherichia coli 5S RNA has three structural domains: implications for processing. Nucleic Acids Research (1988) 16: 7457–7476.
[2] Hartvig L, Christiansen J. Intrinsic termination of T7 RNA polymerase mediated by either RNA or DNA. The EMBO journal (1996) 15: 4767–4774.
[3] Kwon YS, Kang C. Bipartite modular structure of intrinsic, RNA hairpin-independent termination signal for phage RNA polymerases. The Journal of Biological Chemistry (1999) 274: 29149–29155.
[4] Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. Journal of Molecular Biology (1983) 166: 477–535.
[5] Dunn JJ, Studier FW. The transcription termination site at the end of the early region of bacteriophage T7 DNA. Nucleic Acids Research (1980) 8: 2119–2132.