Difference between revisions of "Part:BBa K592012"
(→GreatBay_SCIE 2022's Characterisation) |
|||
| (54 intermediate revisions by 10 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K592012 short</partinfo> | <partinfo>BBa_K592012 short</partinfo> | ||
| − | This chromoprotein eforRed naturally exhibits red color when expressed. The color is weaker than RFP | + | This chromoprotein eforRed naturally exhibits red/pink color when expressed. The color is slightly weaker than RFP. On agar plates and in liquid culture, the color is readily visible to naked eye in less than 24 hours of incubation. The DNA was codon-optimized for expression in E.coli and synthesized by the Korean company Bioneer Corporation. |
| + | '''Important: This part is not available in the registry yet, however, the same part is available from the registry with the RBS B0032: <partinfo>BBa_K1073023</partinfo>.''' | ||
| + | ===Usage and Biology=== | ||
| + | This part is useful as a reporter. | ||
| + | |||
| + | Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989049", https://parts.igem.org/Part:BBa_K1989050" or https://parts.igem.org/Part:BBa_K1989051". | ||
[[Image:eforred_plate_small.jpg|420px]] | [[Image:eforred_plate_small.jpg|420px]] | ||
[[Image:UUChromo.jpg|500px]] | [[Image:UUChromo.jpg|500px]] | ||
| + | '''iGEM12_Uppsala_University:''' ''Left picture:'' Visual color expression of eforRed (<partinfo>BBa_K592012</partinfo>). <br> | ||
| + | ''Right picture:'' The Uppsala chromoprotein collection and RFP. The image shows pellets of ''E coli'' expressing chromoproteins eforRed <partinfo>BBa_K592012</partinfo>, RFP <partinfo>BBa_E1010</partinfo>, cjBlue <partinfo>BBa_K592011</partinfo>, aeBlue <partinfo>BBa_K864401</partinfo>, amilGFP <partinfo>BBa_K592010</partinfo> and amilCP <partinfo>BBa_K592009</partinfo>. | ||
| + | |||
| + | |||
| + | [[Image:eforRed.JPG|420px]]<br> | ||
| + | '''iGEM17_SJTU_BioX_Shanghai:''' | ||
| + | |||
| + | '''J23119+Target3+eforRed '''<partinfo>BBa_K2285012</partinfo><br> | ||
| + | '''J23119+Target1+eforRed ''' <partinfo>BBa_K2285016</partinfo><br> | ||
| + | |||
| + | An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP <partinfo>BBa_K515005</partinfo> and terminator <partinfo>BBa_B1006</partinfo> by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of eforRed expression. | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. ''Diversity and evolution of coral fluorescent proteins.'' PLoS One 3:e2680. | ||
| + | |||
| + | |||
| + | |||
| + | ==GreatBay_SCIE 2022's Characterisation== | ||
| + | |||
| + | ===eforRED as A Reporter Protein=== | ||
| + | |||
| + | Our iGEM project involves the proving of the functionality of our aritificially designed cellulosome complex, which consists of multiple scaffoldins and cell surface-display system cross-linked via dockerin-cohesin interactions and nanobody-antigen interactions, both are strong noncovalent protein-protein interactions(PPIs). To prove the high-affinity interaction between the modules, we designed three eforRED fusion proteins (Fig. 1D). | ||
| + | |||
| + | The nanobody-antigen interaction was verified by mixing intact <i>E.coli</i> cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 1A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 2C). | ||
| + | |||
| + | After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II dockerin fused with eforRED (Fig. 1B). A negative control lacking OlpB-Ag3 was set up for result comparison. Centrifugation was used to remove supernatant and the red fluorescence was only identified in pellets of the sample group, confirming the type II cohesin-dockerin interaction (Fig. 2D). | ||
| + | |||
| + | Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 1C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 2E), verifying the type I cohesin-dockerin interaction. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:GreatBay SCIE--Part Fig5.png|thumbnail|850px|center|'''Figure 1:''' | ||
| + | Cellulosomal scaffold system construct (A) Antigen-nanobody interaction between Nb3 and Ag3 domain reported by the ligated eforRed fluorescent domain (B) Type II cohesin -dockerin interaction between a fixed secondary scaffoldin component and a type II dockerin domain reported by the ligated eforRed fluorescent domain (C) Type I cohesin-dockerin interaction between a fixed primary scaffoldin component and a type I dockerin domain reported by a ligated eforRed fluorescent domain (D) Genetic circuit designed for the expression of ligated form of Ag3, type I dockerin and type II dockerin domains fused with an eforRed fluorescent domain at N terminus for reporting the adhesive functions of those domains.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:GreatBay SCIE--Part Fig6.png|thumbnail|850px|center|'''Figure 2:''' | ||
| + | Production and assay of the scaffold proteins (A) SDS-PAGE analysis for the presence of type I dockerin-eforRed (B) SDS-PAGE analysis of Ag3-eforRed and type II dockerin -eforRed containing type II dockerin fused with an eforRed domain (C) The fluorescence indication for antigen-nanobody interaction for <i>E.coli</i> surface display with an Ag3 control group and a sample group (D) The fluorescence indication for type II cohesin-dockerin interactions with a type II dockerin control group and a sample group (E) The fluorescence indication for type I cohesin-dockerin interactions with a type I dockerin control group and a sample group.]] | ||
| + | |||
| + | ==Jiangsu_High_School 2019’s Characterisation== | ||
| + | |||
| + | ===BBa_K592012 eforRed, red chromoprotein=== | ||
| + | <p>The E. coli expressing red chromoprotein(eforRed) were cultured at 28 °C and the overnight bacterial solution was collected for subsequent measurement.</p> | ||
| + | <p>After ultrasonically disruption of the above bacterial solution ,5 ul of each was added to 4 kinds of 45 ul solution having a pH of 3, 5, 7, and 9, and the fluorescence was measured using varioskan flash of Thermo Scientific. The excitation wavelength/emission wavelength corresponding to each fluorescent protein is: eforRed:555/584.</p> | ||
| + | <p>The results showed that the fluorescent value of each fluorescent protein did not change significantly in acid and alkali circumstance before sonication, but after sonication, each fluorescent protein showed the highest fluorescent value at pH 7, and the fluorescent value decreased as the environment became more acidic.</p> | ||
| + | |||
| + | [[Image:FluorescenceofeforRed.png|400px|center]] | ||
| + | |||
| + | <p>Figure Fluorescence of eforRed under different pH. There were two kinds of eforRed expressing in E.coli(eforRed) and expressing in E.coli after ultrasonically disruption(eforRed -UC). Under the same pH, the fluorescence of eforRed was higher than eforRed -UC. With increasing of pH( among pH3, pH5, pH7,), the fluorescence of eforRed and eforRed -UC all raised. When the pH increased from pH7 to pH9, the fluorescence of eforRed and eforRed were all down.</p> | ||
| + | |||
| + | <p>The results showed that the fluorescent value of each fluorescent protein did not change significantly in acid and alkali circumstance before sonication, but after sonication, each fluorescent protein showed the highest fluorescent value at pH 7, and the fluorescent value decreased as the environment became more acidic.</p> | ||
| + | |||
| + | ==Hong Kong-CUHK 2019’s Characterisation== | ||
| + | |||
| + | <center>[[file:T--Hong Kong-CUHK--IPTG on eforRED summary 1.png]]</center> | ||
| + | <center>[[file:T--Hong_Kong-CUHK--IPTG on eforRED summary 2.png]]</center> | ||
| + | |||
| + | <p> The above figures show the change in color of the <i>E.coli</i> culture in a time-dependent manner under the different IPTG induction. In general, the bacterial pellet shows red color after 3-4 hours of IPTG induction regardless of the IPTG concentration, under the regulation of the lac-T7 promoter. </p> | ||
| + | |||
| + | <center>[[file:T--Hong Kong-CUHK--eforRED mRNA.jpeg]]</center> | ||
| + | |||
| + | <p>The above figure shows the profile of the total eforRED mRNA in the bacterial culture. Expression of eforRed was significantly increased, starting from 6 hours after induction, while it reached maximum at 10 hours after induction and started decline.</p> | ||
| + | |||
| + | ==NJTech_China 2020’s Characterisation== | ||
| + | |||
| + | <p>The eforRed sequence (Part:BBa_K592012) optimized for <i>E. coli</i> was incorporated into <i>E. coli</i> BL21 for protein characterization and data measurement. We conducted a series of experiments to obtain new data of eforRed chromoprotein.</p> | ||
| + | |||
| + | <p>Methods</p> | ||
| + | |||
| + | <p>We performed SDS-PAGE on the eforRed protein to screen the protein expression and detect the effect of purification. Next, we performed the Time of Flight Mass Spectrometer for the eforRed protein sample. We also applied the full wavelength measurement on the eforRed protein. Finally, we used Swiss-Model to simualte the three-dimensional structure of the protein.</p> | ||
| + | |||
| + | <p>Results</p> | ||
| + | |||
| + | [[File: NJTech China eforRed-1.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.1</b> The fermentation broth of eforRed.   <b>Fig.2</b> The cell pellet collected after centrifugation.]] | ||
| + | |||
| + | <p>Conclusion: The cell pellet was collected by harvesting 50mL culture after 24h of induction followed by centrifugation at 6000 rpm for 10 min. Then, we performed ultrasonic disruption and collected the supernatant after centrifugation. The protein was purified and collected through ultrafiltration and affinity chromatography.</p> | ||
| + | |||
| + | [[File: NJTech China eforRed-2.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.3</b> SDS-PAGE of the chromoprotein eforRed]] | ||
| + | |||
| + | <p>1. eforRed- The culture after IPTG induction.</p> | ||
| + | <p>2. eforRed- The culture without IPTG induction.</p> | ||
| + | <p>3. eforRed- Supernatant after IPTG induction and sonication.</p> | ||
| + | <p>4. eforRed- Supernatant sample without IPTG induction after sonication.</p> | ||
| + | <p>5. eforRed- The pellet after IPTG induction and ultrasound.</p> | ||
| + | <p>6. eforRed- The pellet without IPTG induction after ultrasound.</p> | ||
| + | <p>7. eforRed- Protein sample before affinity chromatography.</p> | ||
| + | <p>8. eforRed- Purified protein sample.</p> | ||
| + | <p>9. eforRed- Protein sample after concentration. </p> | ||
| + | <p>Conclusion : The protein gel proved that the molecular mass of the protein was correct (the molecular mass of eforRed protein is about 27kDa), and more eforRed protein can be obtained with IPTG induction (compare lanes 1, 3, 5 with lanes 2, 4, and 6). The protein purification after nickel affinity chromatography was effective (lane 8). The impurity protein was less than the sample before affinity chromatography (lanes 7, 8).</p> | ||
| + | |||
| + | [[File: NJTech China eforRed-3.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.4</b> TOF MS of eforRed]] | ||
| + | |||
| + | <p>Conclusion: We performed TOF-Mass Spectrometry on the purified eforRed chromoprotein. The predicted molecular mass of this protein is about 27kDa, and the exact size is 27306.3Da (the value of the highest peak is shown as the molecular weight of eforRed protein). The other lines are impurity proteins, which may because the nickel affinity chromatography did not remove all impurity proteins or the eforRed protein itself degraded into small fragments.</p> | ||
| + | |||
| + | [[File: NJTech China eforRed-4.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.5</b> Absorption spectrum of eforRed (190 to 1100nm)]] | ||
| + | <p>Conclusion : eforRed protein full-wavelength scan profile :</p> | ||
| + | <p>198nm:2.625A  210nm:2.775A</p> | ||
| + | <p>216nm:2.780A  224nm:2.812A</p> | ||
| + | <p>278nm:0.283A  344nm:0.088A</p> | ||
| + | <p>582nm:0.630A  1066nm:0.017A</p> | ||
| + | <p>The above figure showed the absorption spectrum of eforRed (190 to 1100nm). The full-wavelength scan of eforRed protein showed that the strongest absorption peak of eforRed protein occurs at 224nm.</p> | ||
| + | |||
| + | <p>Structural modeling results of the eforRed protein based on Swiss-Model</p> | ||
| + | [[File: NJTech China eforRed-5.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.6-1</b> The results of the homology and structural modelling protein eforRed]] | ||
| + | [[File: NJTech China eforRed-6.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.6-2</b> The 3D model of the homology and structural modelling protein eforRed]] | ||
| + | |||
| + | <p>Conclusion: We used Swiss-Model to simulate the three-dimensional structure of eforRed protein. The above figures showed the modeling result of Swiss-Model.</p> | ||
| + | |||
| + | ==NDNF_China 2021’s Characterisation== | ||
| + | <html> | ||
| + | |||
| + | <p>In summary: we have tested this genetic part in the Hidro system and we found that this genetic part could be well implemented in the Hidro system for eforRed expression induced by L-arabinose. The result showed that eforRed is a stable protein working in a hydrogel environment. Below is the detailed description.</p> | ||
| + | <p>To determine whether encapsulated bacteria in Hidro can respond to these external signal inputs, we encapsulated Hidro with the bacteria containing a genetic circuit (<a href="https://parts.igem.org/Part:BBa_K3886026">BBa_K3886026</a>) that expresses chromoprotein (eforRed) in response to L-arabinose induction (Figure 1A). We then incubated the beads at 37 °C in the absence of L-arabinose or in the presence of 10mM L-arabinose. We found that encapsulated cells exposed to L-arabinose exhibited a pink colour compared with encapsulated cells not exposed to L-arabinose (Figure 1B). Thus, gene expression in Hidro can be exogenously controlled by chemical inducers. For more information, please visit: <a href="https://2021.igem.org/Team:NDNF_China/Proof_Of_Concept">NDNF_China Proof-Of-Concept</a>.</p> | ||
| + | <div style="text-align: center;"> | ||
| + | <img src="https://2021.igem.org/wiki/images/e/ea/T--NDNF_China--pocf26.png" alt="" width=600"> | ||
| + | <h6 style="text-align:center">Figure 1: (A) The schematic of a L-arabinose–controlled genetic switch to produce chromoprotein eforRed; (B) eforRed protein production in Hidro.</h6> | ||
| + | </div> | ||
| + | </html> | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
| Line 21: | Line 144: | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K592012 parameters</partinfo> | <partinfo>BBa_K592012 parameters</partinfo> | ||
| − | |||
Latest revision as of 00:56, 14 October 2022
eforRed, red chromoprotein
This chromoprotein eforRed naturally exhibits red/pink color when expressed. The color is slightly weaker than RFP. On agar plates and in liquid culture, the color is readily visible to naked eye in less than 24 hours of incubation. The DNA was codon-optimized for expression in E.coli and synthesized by the Korean company Bioneer Corporation.
Important: This part is not available in the registry yet, however, the same part is available from the registry with the RBS B0032: BBa_K1073023.
Usage and Biology
This part is useful as a reporter.
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989049", https://parts.igem.org/Part:BBa_K1989050" or https://parts.igem.org/Part:BBa_K1989051".
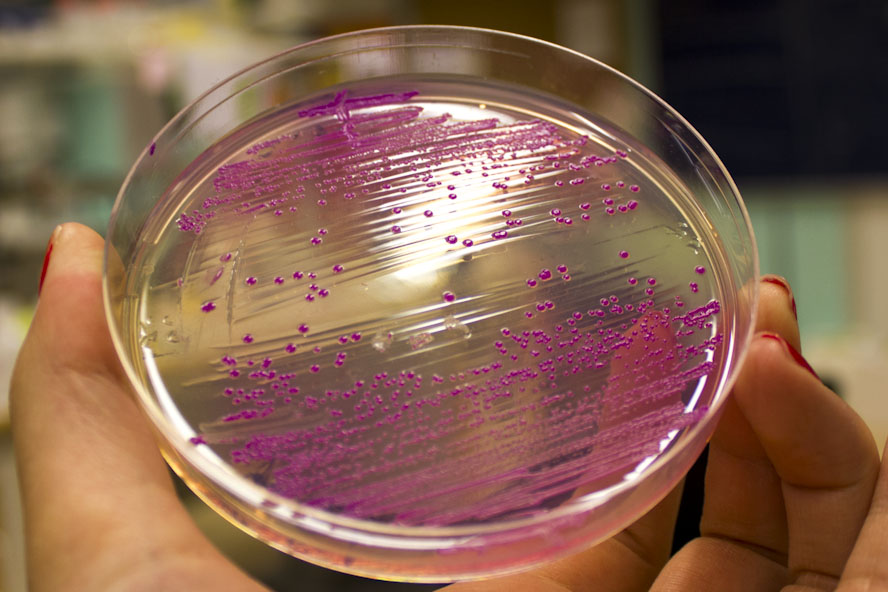
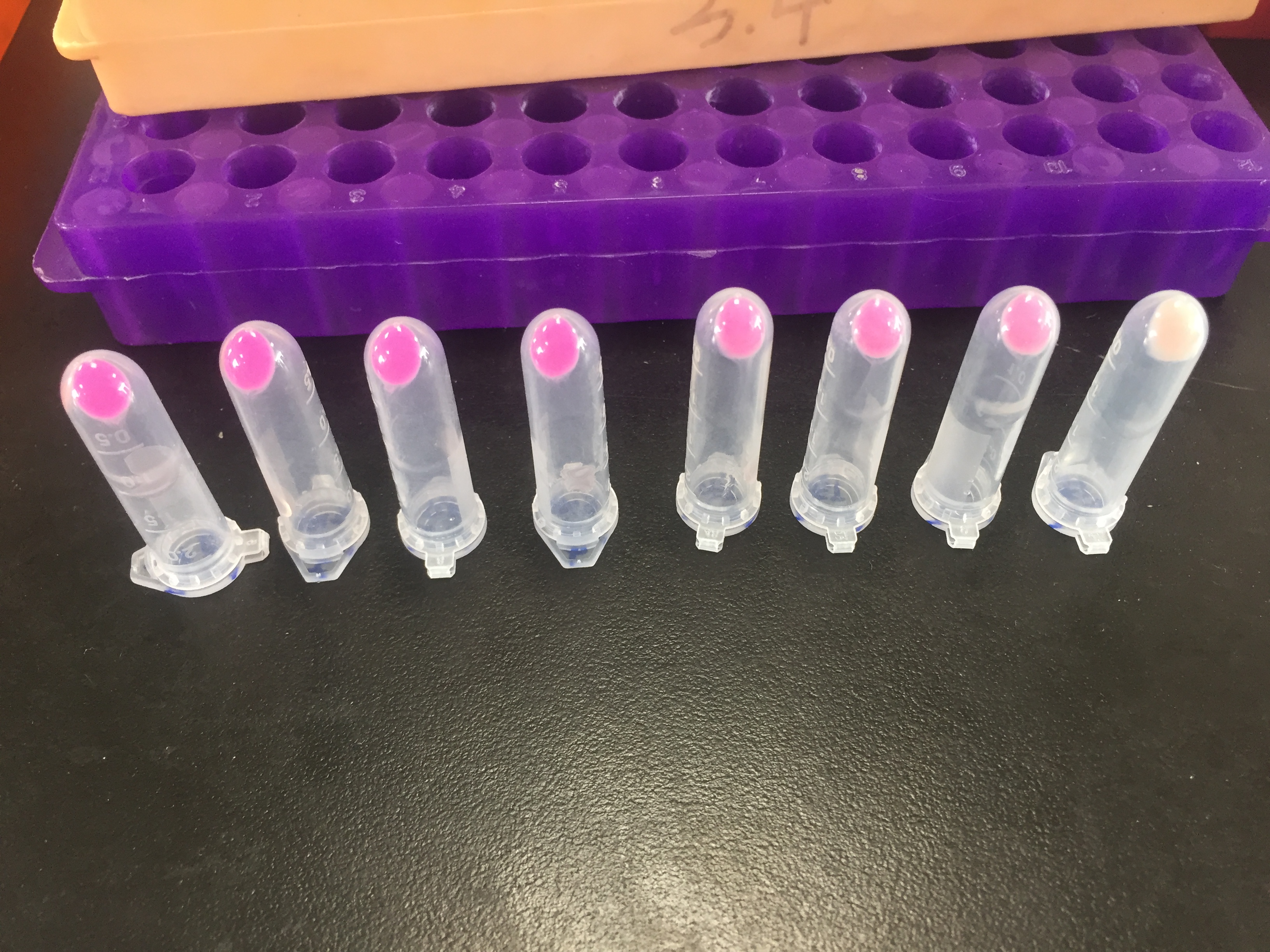
 iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
Right picture: The Uppsala chromoprotein collection and RFP. The image shows pellets of E coli expressing chromoproteins eforRed BBa_K592012, RFP BBa_E1010, cjBlue BBa_K592011, aeBlue BBa_K864401, amilGFP BBa_K592010 and amilCP BBa_K592009.
J23119+Target3+eforRed BBa_K2285012
J23119+Target1+eforRed BBa_K2285016
An improved part has been constructed. Since this part is a coding sequence, we added a RBS which is on the upstream of sfGFP BBa_K515005 and terminator BBa_B1006 by PCR. What's more, our device have a stem-loop structure(called Target) following the constitutive promoter J23119, to achieve further control of eforRed expression.
References
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 3:e2680.
GreatBay_SCIE 2022's Characterisation
eforRED as A Reporter Protein
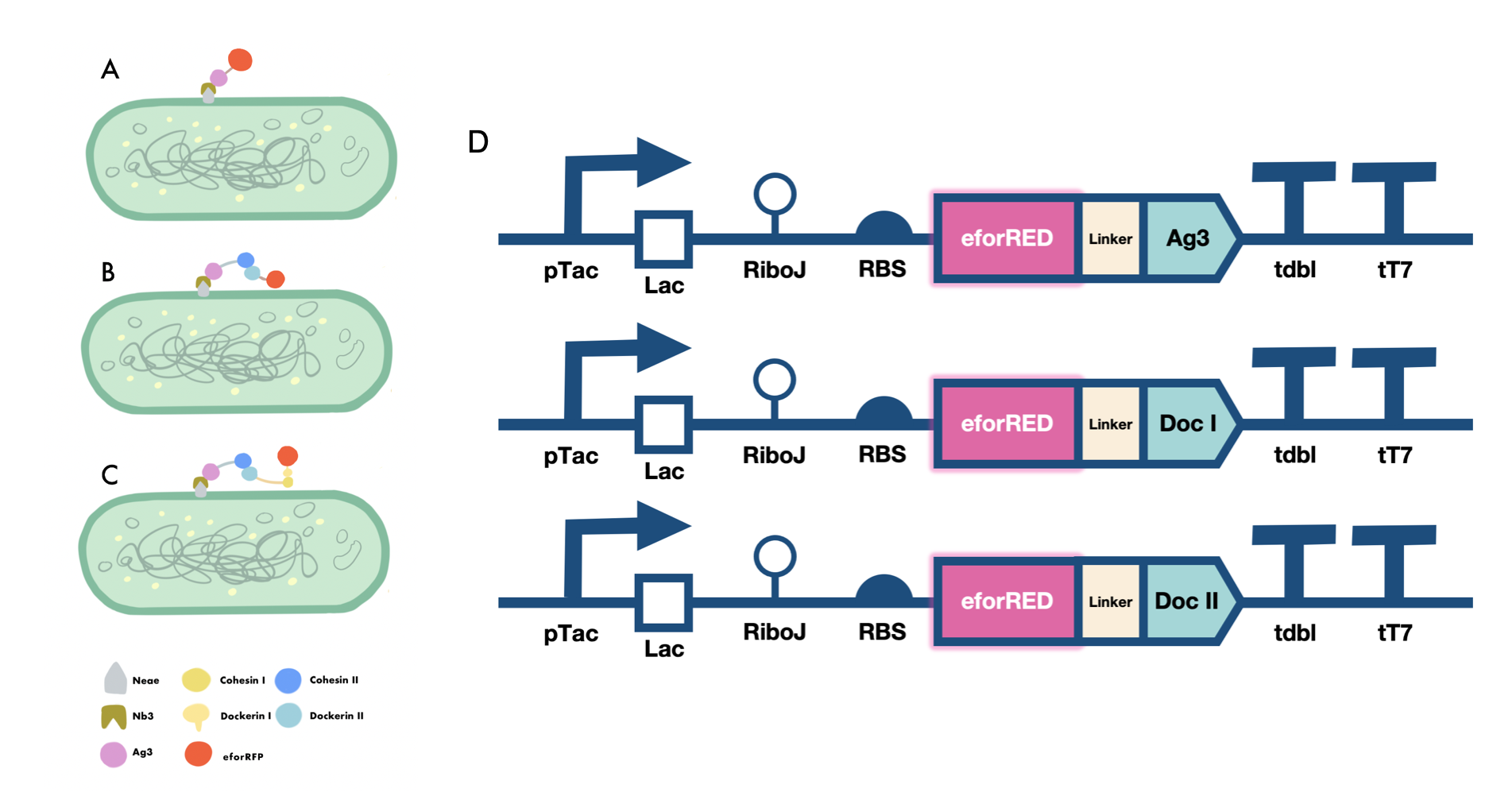
Our iGEM project involves the proving of the functionality of our aritificially designed cellulosome complex, which consists of multiple scaffoldins and cell surface-display system cross-linked via dockerin-cohesin interactions and nanobody-antigen interactions, both are strong noncovalent protein-protein interactions(PPIs). To prove the high-affinity interaction between the modules, we designed three eforRED fusion proteins (Fig. 1D).
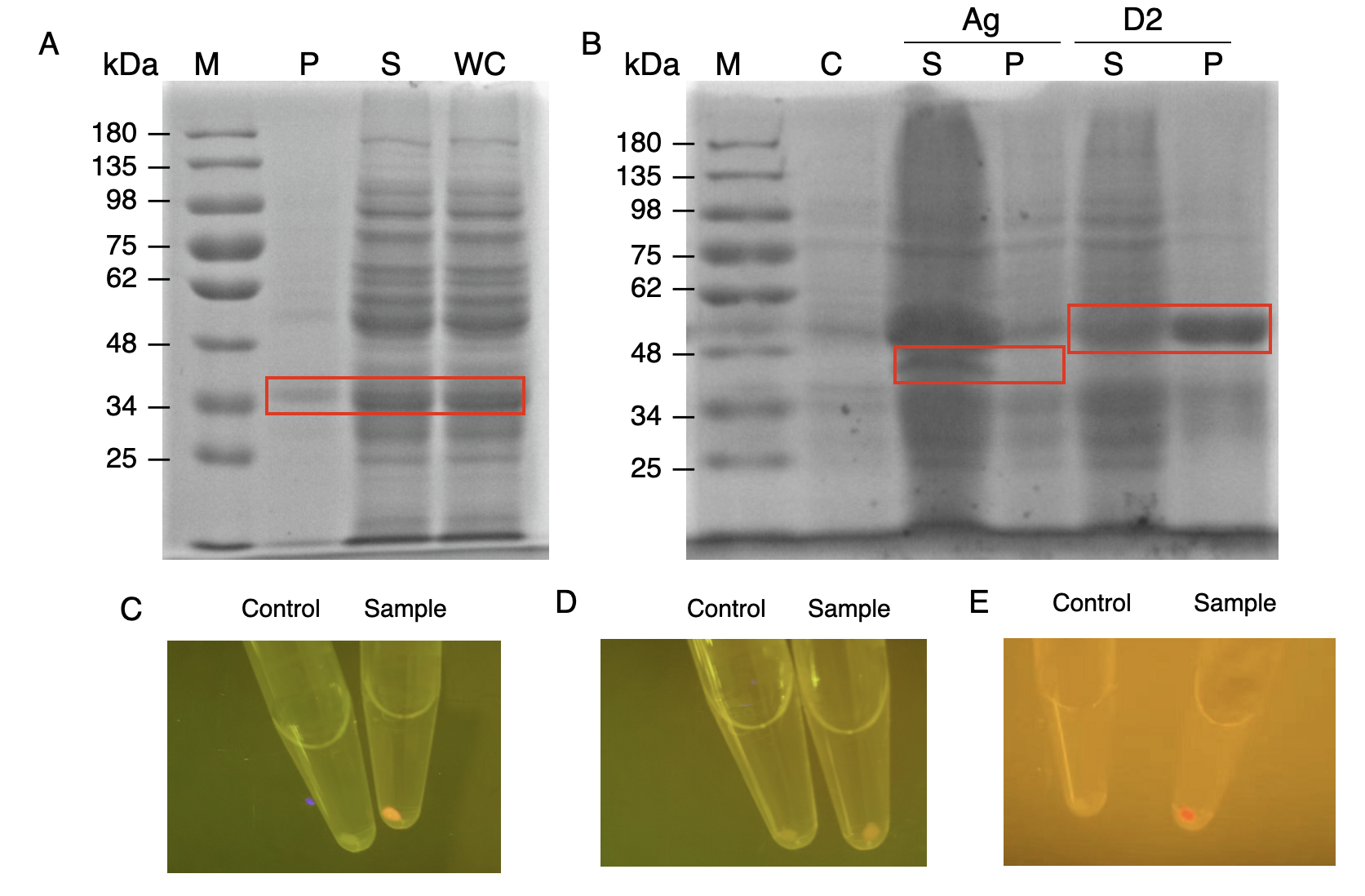
The nanobody-antigen interaction was verified by mixing intact E.coli cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 1A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 2C).
After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II dockerin fused with eforRED (Fig. 1B). A negative control lacking OlpB-Ag3 was set up for result comparison. Centrifugation was used to remove supernatant and the red fluorescence was only identified in pellets of the sample group, confirming the type II cohesin-dockerin interaction (Fig. 2D).
Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 1C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 2E), verifying the type I cohesin-dockerin interaction.
Jiangsu_High_School 2019’s Characterisation
BBa_K592012 eforRed, red chromoprotein
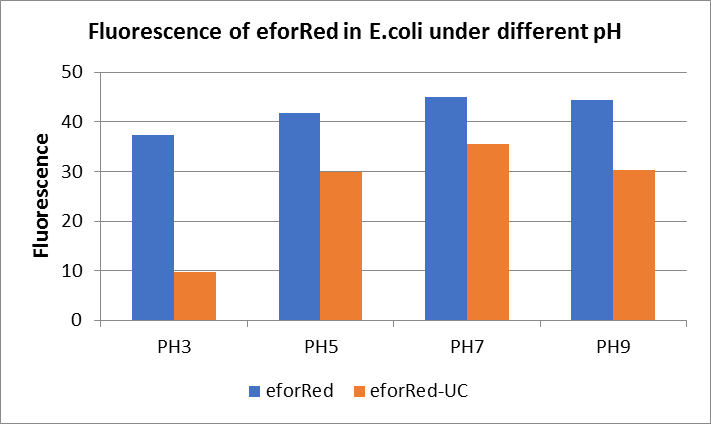
The E. coli expressing red chromoprotein(eforRed) were cultured at 28 °C and the overnight bacterial solution was collected for subsequent measurement.
After ultrasonically disruption of the above bacterial solution ,5 ul of each was added to 4 kinds of 45 ul solution having a pH of 3, 5, 7, and 9, and the fluorescence was measured using varioskan flash of Thermo Scientific. The excitation wavelength/emission wavelength corresponding to each fluorescent protein is: eforRed:555/584.
The results showed that the fluorescent value of each fluorescent protein did not change significantly in acid and alkali circumstance before sonication, but after sonication, each fluorescent protein showed the highest fluorescent value at pH 7, and the fluorescent value decreased as the environment became more acidic.
Figure Fluorescence of eforRed under different pH. There were two kinds of eforRed expressing in E.coli(eforRed) and expressing in E.coli after ultrasonically disruption(eforRed -UC). Under the same pH, the fluorescence of eforRed was higher than eforRed -UC. With increasing of pH( among pH3, pH5, pH7,), the fluorescence of eforRed and eforRed -UC all raised. When the pH increased from pH7 to pH9, the fluorescence of eforRed and eforRed were all down.
The results showed that the fluorescent value of each fluorescent protein did not change significantly in acid and alkali circumstance before sonication, but after sonication, each fluorescent protein showed the highest fluorescent value at pH 7, and the fluorescent value decreased as the environment became more acidic.
Hong Kong-CUHK 2019’s Characterisation
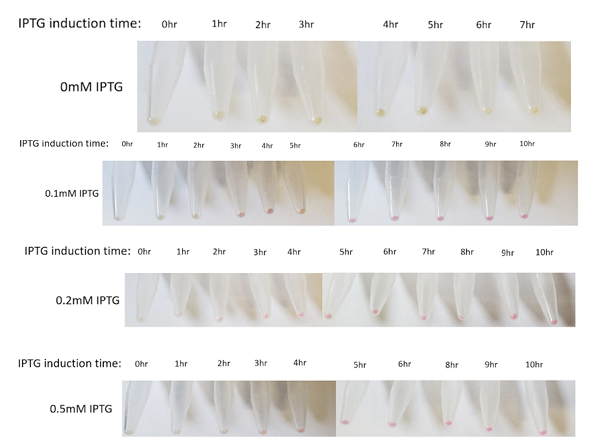

The above figures show the change in color of the E.coli culture in a time-dependent manner under the different IPTG induction. In general, the bacterial pellet shows red color after 3-4 hours of IPTG induction regardless of the IPTG concentration, under the regulation of the lac-T7 promoter.
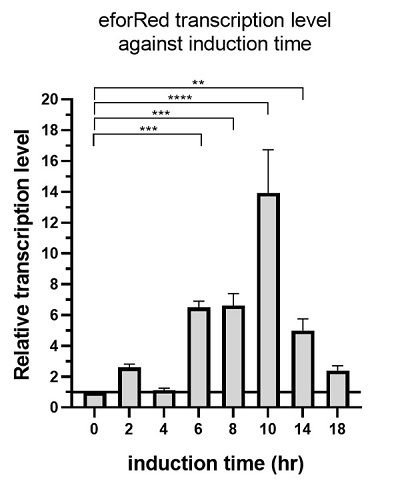
The above figure shows the profile of the total eforRED mRNA in the bacterial culture. Expression of eforRed was significantly increased, starting from 6 hours after induction, while it reached maximum at 10 hours after induction and started decline.
NJTech_China 2020’s Characterisation
The eforRed sequence (Part:BBa_K592012) optimized for E. coli was incorporated into E. coli BL21 for protein characterization and data measurement. We conducted a series of experiments to obtain new data of eforRed chromoprotein.
Methods
We performed SDS-PAGE on the eforRed protein to screen the protein expression and detect the effect of purification. Next, we performed the Time of Flight Mass Spectrometer for the eforRed protein sample. We also applied the full wavelength measurement on the eforRed protein. Finally, we used Swiss-Model to simualte the three-dimensional structure of the protein.
Results
Conclusion: The cell pellet was collected by harvesting 50mL culture after 24h of induction followed by centrifugation at 6000 rpm for 10 min. Then, we performed ultrasonic disruption and collected the supernatant after centrifugation. The protein was purified and collected through ultrafiltration and affinity chromatography.
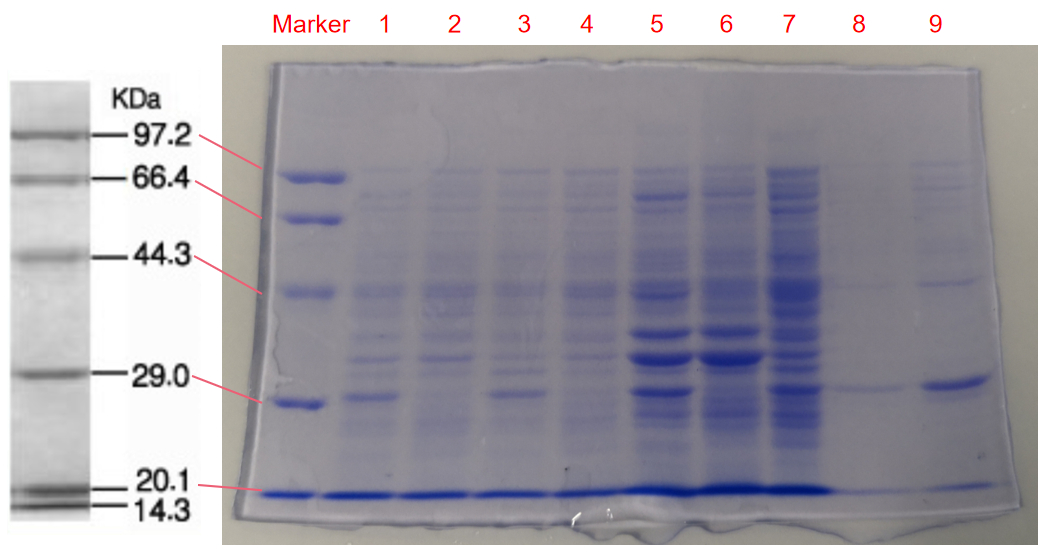
1. eforRed- The culture after IPTG induction.
2. eforRed- The culture without IPTG induction.
3. eforRed- Supernatant after IPTG induction and sonication.
4. eforRed- Supernatant sample without IPTG induction after sonication.
5. eforRed- The pellet after IPTG induction and ultrasound.
6. eforRed- The pellet without IPTG induction after ultrasound.
7. eforRed- Protein sample before affinity chromatography.
8. eforRed- Purified protein sample.
9. eforRed- Protein sample after concentration.
Conclusion : The protein gel proved that the molecular mass of the protein was correct (the molecular mass of eforRed protein is about 27kDa), and more eforRed protein can be obtained with IPTG induction (compare lanes 1, 3, 5 with lanes 2, 4, and 6). The protein purification after nickel affinity chromatography was effective (lane 8). The impurity protein was less than the sample before affinity chromatography (lanes 7, 8).
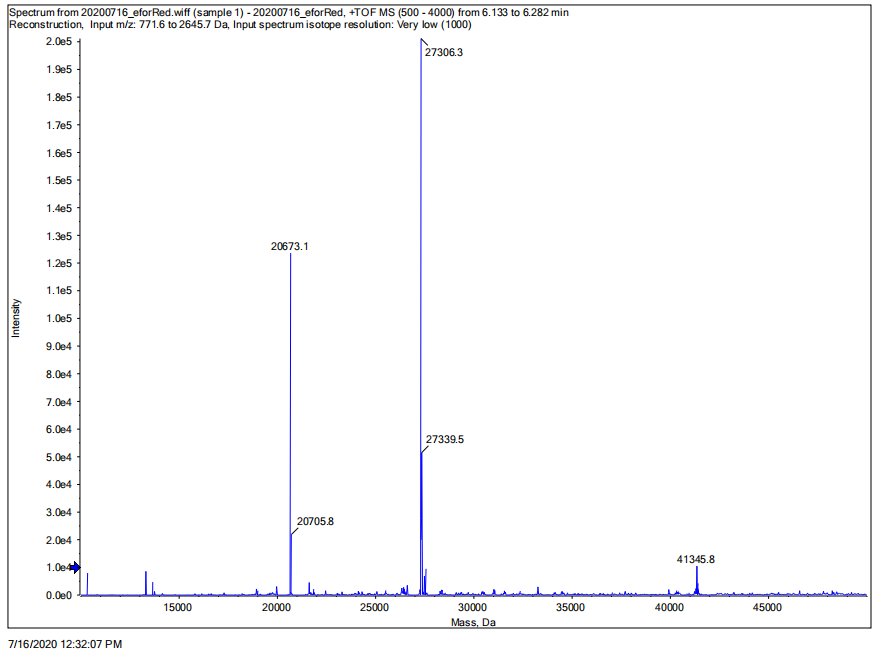
Conclusion: We performed TOF-Mass Spectrometry on the purified eforRed chromoprotein. The predicted molecular mass of this protein is about 27kDa, and the exact size is 27306.3Da (the value of the highest peak is shown as the molecular weight of eforRed protein). The other lines are impurity proteins, which may because the nickel affinity chromatography did not remove all impurity proteins or the eforRed protein itself degraded into small fragments.
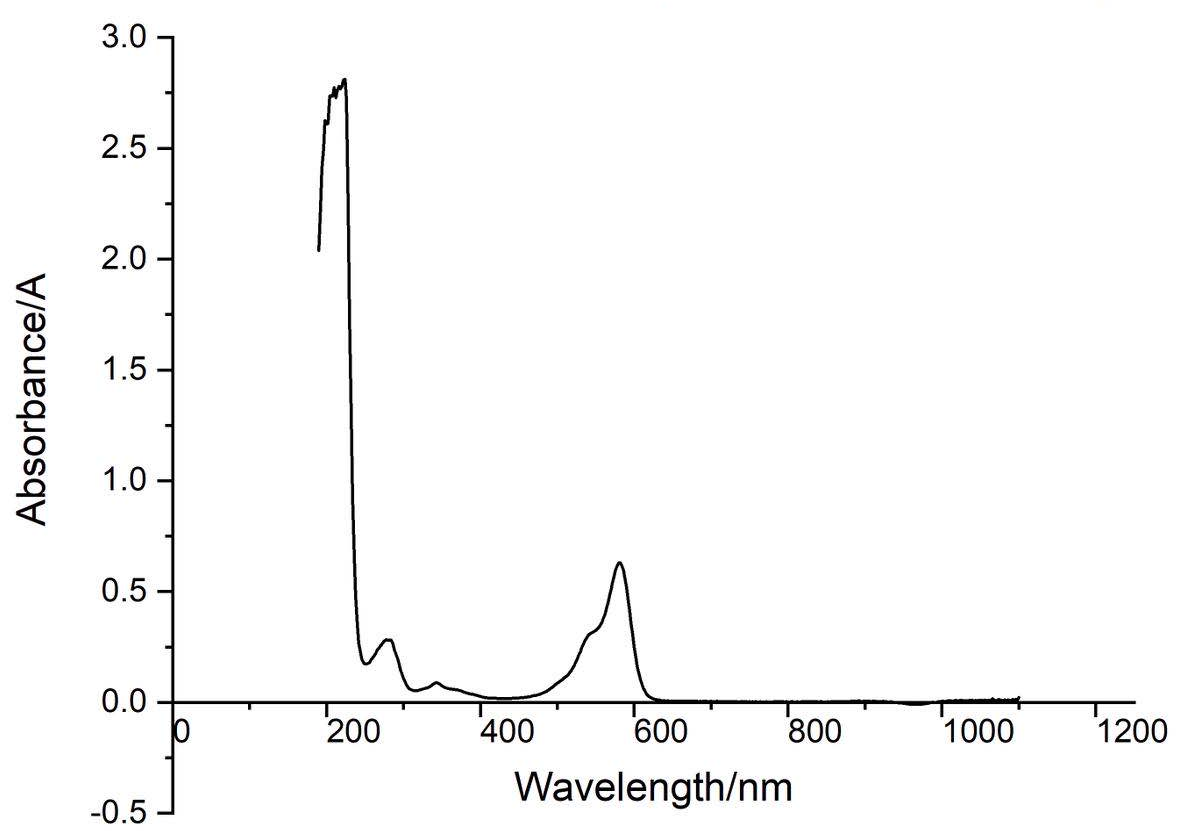
Conclusion : eforRed protein full-wavelength scan profile :
198nm:2.625A 210nm:2.775A
216nm:2.780A 224nm:2.812A
278nm:0.283A 344nm:0.088A
582nm:0.630A 1066nm:0.017A
The above figure showed the absorption spectrum of eforRed (190 to 1100nm). The full-wavelength scan of eforRed protein showed that the strongest absorption peak of eforRed protein occurs at 224nm.
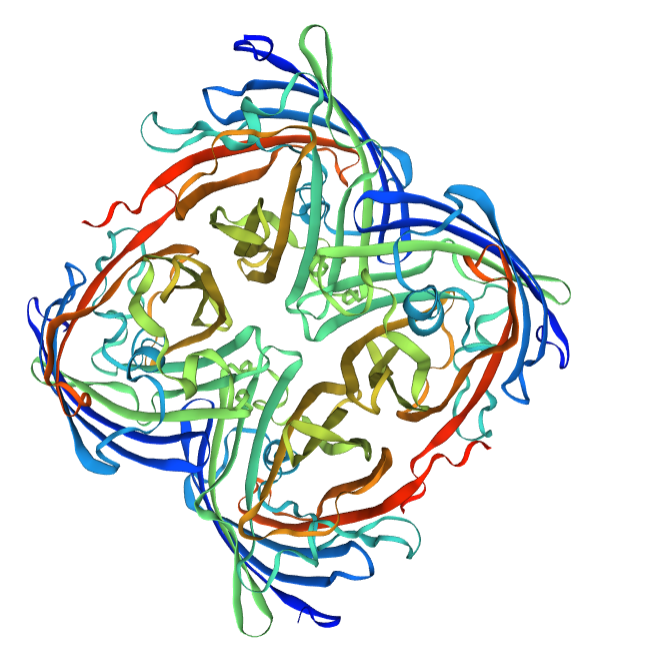
Structural modeling results of the eforRed protein based on Swiss-Model
Conclusion: We used Swiss-Model to simulate the three-dimensional structure of eforRed protein. The above figures showed the modeling result of Swiss-Model.
NDNF_China 2021’s Characterisation
In summary: we have tested this genetic part in the Hidro system and we found that this genetic part could be well implemented in the Hidro system for eforRed expression induced by L-arabinose. The result showed that eforRed is a stable protein working in a hydrogel environment. Below is the detailed description.
To determine whether encapsulated bacteria in Hidro can respond to these external signal inputs, we encapsulated Hidro with the bacteria containing a genetic circuit (BBa_K3886026) that expresses chromoprotein (eforRed) in response to L-arabinose induction (Figure 1A). We then incubated the beads at 37 °C in the absence of L-arabinose or in the presence of 10mM L-arabinose. We found that encapsulated cells exposed to L-arabinose exhibited a pink colour compared with encapsulated cells not exposed to L-arabinose (Figure 1B). Thus, gene expression in Hidro can be exogenously controlled by chemical inducers. For more information, please visit: NDNF_China Proof-Of-Concept.
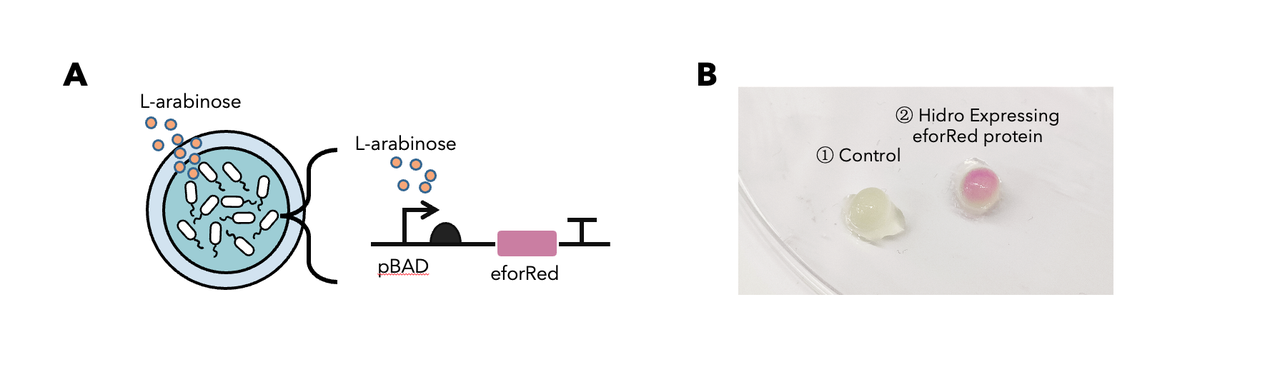
Figure 1: (A) The schematic of a L-arabinose–controlled genetic switch to produce chromoprotein eforRed; (B) eforRed protein production in Hidro.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]