Difference between revisions of "Part:BBa K733012"
| (7 intermediate revisions by 3 users not shown) | |||
| Line 10: | Line 10: | ||
==Characterization== | ==Characterization== | ||
| + | |||
| + | '''Background Information''' ([http://2012.igem.org/Team:HKUST-Hong_Kong/Module/Regulation_and_control link to Regulation and Control Module]) | ||
| + | |||
| + | The rationale for including this growth inhibition device is that over-dose of BMP2 can cause unexpected proliferation of normal colon cells.(Zhang et al., 2012) Thus, a growth inhibition device is introduced and will be characterized. | ||
| + | |||
'''Objective''' | '''Objective''' | ||
| − | The objective of this characterization is to find out what is the minimal concentration of xylose to inhibit the growth of our | + | The objective of this characterization is to find out what is the minimal concentration of xylose necessary to inhibit the growth of our B. hercules. |
| Line 20: | Line 25: | ||
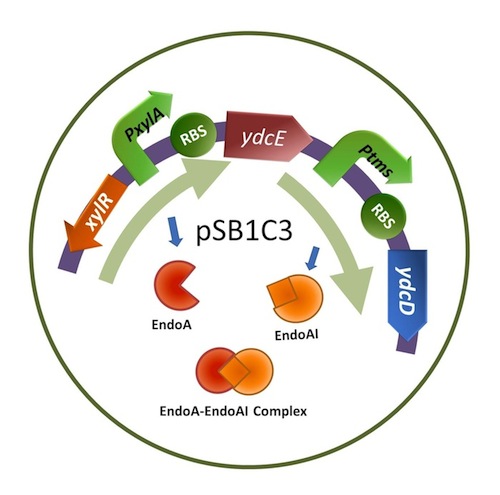
''I. Construct.'' | ''I. Construct.'' | ||
| − | [[Image: | + | [[Image:CGIDchar smaller.JPG]] |
| − | [https://parts.igem.org/Part:BBa_K733002 xylR]: The transcriptional regulator for the xylose inducible promoter. | + | [https://parts.igem.org/Part:BBa_K733002 ''xylR'']: The transcriptional regulator for the xylose inducible promoter. |
| − | [https://parts.igem.org/Part:BBa_K733002 PxylA]: The xylose inducible promoter. | + | [https://parts.igem.org/Part:BBa_K733002 ''PxylA'']: The xylose inducible promoter. |
| − | [https://parts.igem.org/Part:BBa_K733004 ydcE] (ndoA): The toxin gene encoding EndoA. | + | [https://parts.igem.org/Part:BBa_K733004 ''ydcE''] (''ndoA''): The toxin gene encoding EndoA. |
| − | [https://parts.igem.org/Part:BBa_K733001 | + | [https://parts.igem.org/Part:BBa_K733001 ''Ptms'']: The low efficient constitutive promoter. |
| − | [https://parts.igem.org/Part:BBa_K733003 ydcD] (endB): The antitoxin gene encoding YdcD. | + | [https://parts.igem.org/Part:BBa_K733003 ''ydcD''] (''endB''): The antitoxin gene encoding YdcD (EndoAI). |
''II. Culture Medium.'' | ''II. Culture Medium.'' | ||
| − | Supplemented M9 minimal medium (M9 salt, 1 mM thiamine hydrochloride, 0.2% casamino acids, 0.1 M | + | Supplemented M9 minimal medium (M9 salt, 1 mM thiamine hydrochloride, 0.2% casamino acids, 0.1 M MgSO<sub>4</sub>, 0.5 M CaCl<sub>2</sub>, 0.4% glycerol) was used for our characterization. The reason for using this medium with 0.4% glycerol as the carbon source is that glucose can repress the induction of xylose. (Kim, Mogk & Schumann,. 1996) 25 mg/mL chloramphenicol was diluted 100 times and added to the medium to select the bacteria with our intended vectors. The final concentration gradient of xylose in the supplemented M9 minimal medium is: 0.05%, 0.10%, 0.15%, 0.20% and 0.25%. |
''III. Control and Experiment Group.'' | ''III. Control and Experiment Group.'' | ||
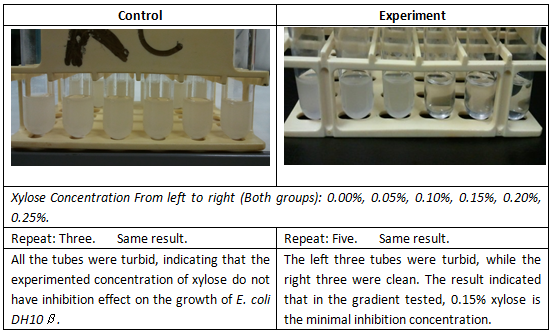
| − | Control Group: | + | Control Group: E. coli DH10B without any vector was used as the control. It was inoculated in the supplemented M9 minimal medium with xylose concentrations: 0.00%, 0.05%, 0.10%, 0.15%, 0.20%, 0.25%. Note that in the control group, the medium does not contain chloramphenicol. |
| − | Experiment Group: | + | Experiment Group: E. coli DH10B with our cell growth inhibition device were inoculated in the supplemented M9 minimal medium with xylose concentrations: 0.00%, 0.05%, 0.10%, 0.15%, 0.20%, 0.25%. The total volume of the culture was 2mL for each test tube. |
''IV. Experiment.'' | ''IV. Experiment.'' | ||
| − | The bacteria cultures were incubated | + | The bacteria cultures were incubated at 37 degrees Celsius and shaken at 200 rpm for exactly 16 hours. After 16-hours incubation, the turbidity of each culture was checked and photographed. Later, 50uL of culture from one set of the experiments was aliquoted and spread on an LB plate containing chloramphenicol (25 ug/mL) for overnight incubation. |
| − | + | ||
| Line 55: | Line 59: | ||
[[Image:CGIDC.png]] | [[Image:CGIDC.png]] | ||
| − | ''Note that after spreading the culture on the plates, there were bacteria growing for all the tubes in the experiment groups. Admittedly, there were distinguishable | + | ''Note that after spreading the culture on the plates, there were bacteria growing for all the tubes in the experiment groups. Admittedly, there were distinguishable differences between the concentrations of 0.00%, 0.05% and 0.10% and the concentration of 0.15%, 0.20% and 0.25%: for the left three groups, the number of bacteria was much more than that in the right three ones. This result indicated that our E. coli had its growth inhibited and did not die from the xylose induced toxin expression.'' |
| + | |||
| + | |||
| + | |||
| + | '''References''' | ||
| + | |||
| + | Pellegrini O, Mathy N, Gogos A, Shapiro L, and Condon C. "The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor.." Molecular microbiology. 56.5 (2005): 1139-1148. Print. | ||
| + | Kim, L., Mogk, A., & Schumann, W. (1996). A xylose-inducible Bacillus subtilis integration vector and its application.. Gene, 181(1-2), 71-76. | ||
| + | Zhang J, Ge Y, Sun L, Cao J, Wu Q, Guo L, Wang Z. Effect of Bone Morphogenetic Protein-2 on Proliferation and Apoptosis of Gastric Cancer Cells. Int J Med Sci 2012; 9(2):184-192. | ||
Latest revision as of 23:26, 26 September 2012
xylR+PxylA+RBS+ydcE+Ptms+RBS+ydcD: Cell Growth Inhibition Device
This construct is for the characterization of the cell growth inhibition device. pTms+RBS+ydcD is for the stabilization of the cell growth inhibition system. By this stabilizer, under low level of induction, YdcD will counteract EndoA to protect cell from being sick. (Pellegrini et al., 2005) However, as long as the induction level is strong enough and reaches a threshold, the amount of EndoA accumulated in the cell can inhibit the growth of the bacteria.
Ptms+RBS+ydcD: BBa_K733010
Xylose inducible promoter with ydcE gene: BBa_K733011
Characterization
Background Information ([http://2012.igem.org/Team:HKUST-Hong_Kong/Module/Regulation_and_control link to Regulation and Control Module])
The rationale for including this growth inhibition device is that over-dose of BMP2 can cause unexpected proliferation of normal colon cells.(Zhang et al., 2012) Thus, a growth inhibition device is introduced and will be characterized.
Objective
The objective of this characterization is to find out what is the minimal concentration of xylose necessary to inhibit the growth of our B. hercules.
Method
I. Construct.
xylR: The transcriptional regulator for the xylose inducible promoter.
PxylA: The xylose inducible promoter.
ydcE (ndoA): The toxin gene encoding EndoA.
Ptms: The low efficient constitutive promoter.
ydcD (endB): The antitoxin gene encoding YdcD (EndoAI).
II. Culture Medium.
Supplemented M9 minimal medium (M9 salt, 1 mM thiamine hydrochloride, 0.2% casamino acids, 0.1 M MgSO4, 0.5 M CaCl2, 0.4% glycerol) was used for our characterization. The reason for using this medium with 0.4% glycerol as the carbon source is that glucose can repress the induction of xylose. (Kim, Mogk & Schumann,. 1996) 25 mg/mL chloramphenicol was diluted 100 times and added to the medium to select the bacteria with our intended vectors. The final concentration gradient of xylose in the supplemented M9 minimal medium is: 0.05%, 0.10%, 0.15%, 0.20% and 0.25%.
III. Control and Experiment Group.
Control Group: E. coli DH10B without any vector was used as the control. It was inoculated in the supplemented M9 minimal medium with xylose concentrations: 0.00%, 0.05%, 0.10%, 0.15%, 0.20%, 0.25%. Note that in the control group, the medium does not contain chloramphenicol.
Experiment Group: E. coli DH10B with our cell growth inhibition device were inoculated in the supplemented M9 minimal medium with xylose concentrations: 0.00%, 0.05%, 0.10%, 0.15%, 0.20%, 0.25%. The total volume of the culture was 2mL for each test tube.
IV. Experiment.
The bacteria cultures were incubated at 37 degrees Celsius and shaken at 200 rpm for exactly 16 hours. After 16-hours incubation, the turbidity of each culture was checked and photographed. Later, 50uL of culture from one set of the experiments was aliquoted and spread on an LB plate containing chloramphenicol (25 ug/mL) for overnight incubation.
Result
Note that after spreading the culture on the plates, there were bacteria growing for all the tubes in the experiment groups. Admittedly, there were distinguishable differences between the concentrations of 0.00%, 0.05% and 0.10% and the concentration of 0.15%, 0.20% and 0.25%: for the left three groups, the number of bacteria was much more than that in the right three ones. This result indicated that our E. coli had its growth inhibited and did not die from the xylose induced toxin expression.
References
Pellegrini O, Mathy N, Gogos A, Shapiro L, and Condon C. "The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor.." Molecular microbiology. 56.5 (2005): 1139-1148. Print.
Kim, L., Mogk, A., & Schumann, W. (1996). A xylose-inducible Bacillus subtilis integration vector and its application.. Gene, 181(1-2), 71-76.
Zhang J, Ge Y, Sun L, Cao J, Wu Q, Guo L, Wang Z. Effect of Bone Morphogenetic Protein-2 on Proliferation and Apoptosis of Gastric Cancer Cells. Int J Med Sci 2012; 9(2):184-192.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 847
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Reference
Pellegrini O, Mathy N, Gogos A, Shapiro L, and Condon C. "The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor.." Molecular microbiology. 56.5 (2005): 1139-1148. Print.
Kim, L., Mogk, A., & Schumann, W. (1996). A xylose-inducible Bacillus subtilis integration vector and its application.. Gene, 181(1-2), 71-76.