Difference between revisions of "Part:BBa K907000"
| (27 intermediate revisions by 6 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K907000 short</partinfo> | <partinfo>BBa_K907000 short</partinfo> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <h1> Characterization by ETH Zurich 2016 </h1> | |
| − | '''< | + | <ul> |
| − | < | + | <li>'''Group:''' ETH Zurich 2016 |
| − | < | + | <li>'''Author:''' Asli Azizoglu, Lukas Schmidheini |
| − | + | <li>'''Summary:''' We cloned and characterised a codon optimised Bxb1 for <i>E. coli</i>, and sent it to the registry as | |
| − | + | a biobrick. Our biobrick can be found with a fast degradation tag [[Part:BBa_K2116009]] and here without any degradation | |
| − | + | tag [[Part:BBa_K2116026]]. The non-codon optimised version can be found here [[Part:BBa_K907000]]. Besides some favorable features, | |
| − | + | the codon optimized Bxb1 also supports full assembly compatibility. | |
| + | </ul> | ||
| − | <!-- Add more about the biology of this part here | + | <h1>Biological Characterisation</h1> |
| + | <p>Bxb1 is a phage integrase that mediates unidirectional site-specific recombination between two DNA recognition sequences, the phage attachement | ||
| + | site, 'attP', and the bacterial attachment site, 'attB'. This recombination event is hereon referred to as a flip. Bxb1 belongs to the family of serine integrases and uses a catalytic serine for strand cleavage, recognizes | ||
| + | shorter attP sequences, and does not require host cofactors. It mediates efficient site-specific recombination between two different sequences that are relatively short | ||
| + | yet long enough to be specific on a genomic scale. These properties make Bxb1 an efficient tool for creating genetic logic gates, as demonstrated in various papers. | ||
| + | </p> | ||
| + | |||
| + | <h1>Kinetic Characterisation</h1> | ||
| + | <p>We used a reporter construct [[Part:BBa_K2116024]] with a promoter flanked between attB and attP sites [[Part:BBa_K2116021]]. Thus, we could investigate | ||
| + | the kinetics of Bxb1 using flow cytometry (BD LSR Fortessa SORP) on time course samples. </p> | ||
| + | |||
| + | <h2>Genetic Design</h2> | ||
| + | <ul> | ||
| + | <li> Bxb1 was expressed under a tet promoter, without any degradation tag [[Part:BBa_K2116056]]. The RBS has a strength of 1209.69 au (translation | ||
| + | initiation rate), as calculated by the Salis Lab RBS Calculator[1]. This construct was cloned on a medium | ||
| + | copy plasmid with the p15A replication of origin and Chloramphenicol resistance. | ||
| + | <li> TetR was expressed under the control of medium-strength constitutive promoter [[Part:Bba_J23118]], and cloned onto a | ||
| + | low copy plasmid with pSC101/Rep101 replication of origin and Kanamycine resistance. | ||
| + | <li> We applied a dual fluorescence reporter system [[Part:BBa_K2116024]] that expresses sfGFP [[Part:BBa_K2116017]] when Bxb1 flips a directional | ||
| + | promoter. In the non-flipped state the reporter expresses the red fluorescent protein mNectarine [[Part:BBa_K2116016]]. | ||
| + | </ul> | ||
| + | <h2 id="experimental_setup"> Experimental Setup </h2> | ||
| + | <ul> | ||
| + | <li> The test construct with bxb1 (pBR322/rop replication of origin and Ampiciline resistance) was transformed in <i>E. coli</i> TOP10 together with the reporter construct, and either with TetR or with the empty | ||
| + | backbone as control. | ||
| + | <li> TOP10 cells were grown in LB medium (containing Kanamycine, Ampiciline (50 µg/ml) and Chloramphenicol (34 µg/ml)) for 4 hours, and then | ||
| + | transfered to minimal M9 medium (containing 0.4% glucose, supplemented with L-leucine, L-isoleucine, and L-valine, Kanamycine, Ampiciline (25 µg/ml and Chloramphenicol (17µg/ml)) | ||
| + | with a 1:100 dilution and grown over-night. The culture was again diluted 1:100 in minimal M9 medium the next day. aTc (anhydrotetracycline) was added | ||
| + | at given concentrations at OD<sub>600</sub>0.5. Samples were taken at the specified time points, spun down and resuspended | ||
| + | in PBS at OD<sub>600</sub> 0.01 and kept on ice until measurement. | ||
| + | <li> Results are presented as mean fluorescence per cell and the standard error of the mean. | ||
| + | </ul> | ||
| + | |||
| + | <h2>Results</h2> | ||
| + | <p>We observed that maximum flipping (which we defined as the flipping reached by the used positive controles) is reached at around 11 hours | ||
| + | under these conditions. The response correlates with aTc concentration. | ||
| + | However, compared to the control, where there is no TetR, we see 6-fold less flipping. This could be improved by increasing | ||
| + | the concentration of aTc, however we have found that at 8000 ng/mL cell growth was inhibited.</p> | ||
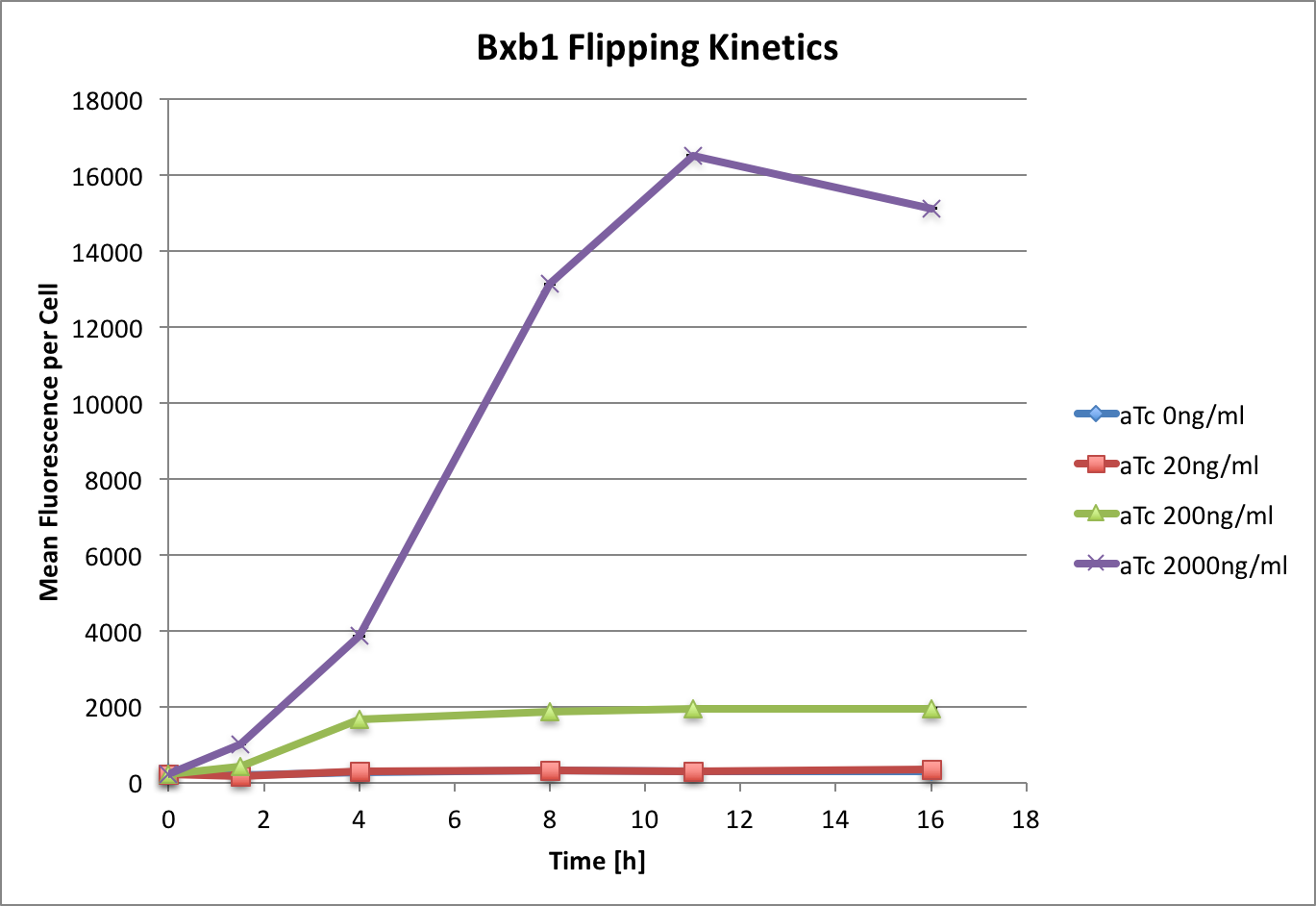
| + | |||
| + | [[File:T--ETH Zurich--codonoptimisation2.png|500px|thumb|center|<b>Figure 1:</b> Time and dose response of bxb1 flipping a directional promoter. | ||
| + | Induction at 0 hours, OD<sub>600</sub> 0.5, with aTc at given concentrations. Data measured by flow cytometry at given time points. | ||
| + | Values are shown as mean fluorescence per cell, error bars indicate SEM. ]] | ||
| + | |||
| + | <h1> Comparison between Codon Optimised and Registry Bxb1 Integrase </h1> | ||
| + | <p>We compared the kinetics of our codon optimised Bxb1 [[Part:BBa_K2116026]] and the one found previously on the registry | ||
| + | [[Part:BBa_K907000]] that was not codon optimized. The same genetic and experimental setup was used as above. The only | ||
| + | difference between the two constructs is that the non-codon optimised bxb1 has a weaker RBS, with a strength of 251.82 au | ||
| + | (translation initiation rate) [1]. Due to this difference | ||
| + | in construction we compared each Bxb1 to its own control (see above, [[#experimental_setup|Experimental Setup]]).</p> | ||
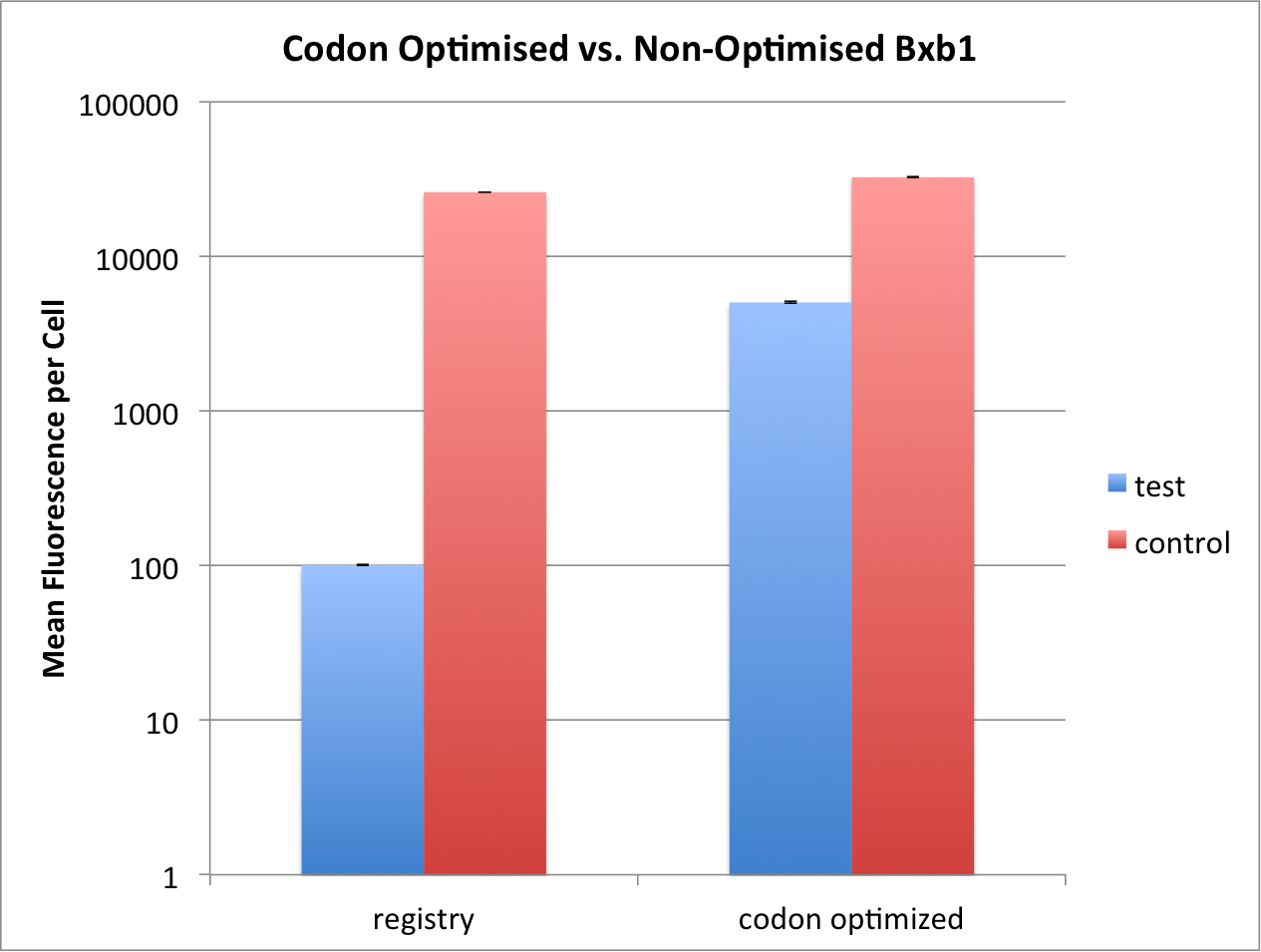
| + | <p> We could show that the codon optimised Bxb1 could reach 85% of maximal flipping, whereas the non-codon optimised | ||
| + | one reached only 0.4% after 16 hours of aTc exposure. We thus argue that the codon optimized Bxb1 seems to be more efficient, however, | ||
| + | the influence of the different RBS strengths was not properly examined.</p> | ||
| + | |||
| + | [[File:T--ETH Zurich--codonoptimisation.png|500px|thumb|center|<b>Figure 2:</b> Comparison of codon optimised and non-optimised Bxb1 flipping | ||
| + | efficiency. Test constructs have Bxb1 expressed under the Tet promoter, with TetR present in the system. Control does not | ||
| + | have TetR in the system, and thus constitutively expresses Bxb1. While non-codon optimised Bxb1 can only reach 0.4% | ||
| + | of the full flipping reached by control, the codon optimised one reaches 85% of its own control. Induction at 0 hours and OD<sub>600</sub> 0.5 with 2000ng/mL. Data measured by flow cytometry at 16h. Values are shown as mean fluorescence per cell, error bars | ||
| + | indicate SEM. ]] | ||
| + | <h2>Improvement</h2> | ||
| + | |||
| + | This part has been improved by iGEM Peking 2017. | ||
| + | The improved Part is submitted as BBa_K2243012;https://parts.igem.org/Part:BBa_K2243012 | ||
| + | |||
| + | =Characterization by Fudan_China 2017= | ||
| + | *'''Group:''' Fudan_China 2017 | ||
| + | *'''Author:''' Haiyun Liu, Fudan University | ||
| + | We characterized the kinetic characterization and the orthogonality between Bxb1 and some other serine recombinase. The attB/P site we used for these experiments were Bxb1 attB ([[Part:BBa_K1039032|BBa_K1039032]]) and attP ([[Part:BBa_K1039031|BBa_K1039031]]) | ||
| + | |||
| + | ==Kinetic Characterization== | ||
| + | We conducted qPCR to record the ratio of attL/R to the sum of attL/R and attB/P, to characterize the efficiency varying with time. | ||
| + | |||
| + | ===Genetic design=== | ||
| + | *The integrase is expressed under a PBad promoter ([[Part:BBa_I13453|BBa_I13453]]). Considering the leakage of PBad promoter would lead to flipping,the promoter is followed by a weaker RBS ([[Part:BBa_B0033|BBa_B0033]]). Ahead of the promoter,an attB and attP are opposite to each other,flanking a 50 bp random sequence. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance. | ||
| + | *To reduce leakage when not induced, AraC is expressed under a constitutive promoter, cloned on a high copy plasmid with pUC19 replication of origin and Ampicillin resistance. | ||
| + | |||
| + | ===Experimental Setup=== | ||
| + | *Since transforming the two plasmid above at a time was unable to prevent the flipping, the plasmid with AraC was first transformed into ''E.coli'' DH10b, then the plasmid with the integrase was transformed into the competent cell with the AraC plasmid. | ||
| + | *After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose for 8 hours. We sampled the culture at 0 hour, the 4th hour and the 8th hour. | ||
| + | *We calculated flipped and unflipped random sequence by qPCR. | ||
| + | |||
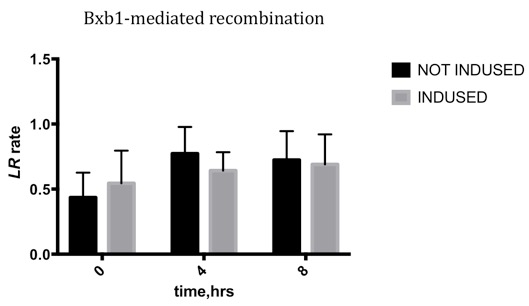
| + | ===Results=== | ||
| + | Due to the leakage before inducing, the LR rate changing is not obvious, which means the Bxb1 integrase is highly efficient, and the leakage may be reduced by changing the genetic design. | ||
| + | |||
| + | [[File:Fudan_China_Bxb1.jpg]] | ||
| + | |||
| + | ==Orthogonality Characterization== | ||
| + | Since our circuit are using multiple integrases, it's important that the attB/P site of one integrase won't be flipped by one another integrase. We conducted orthogonality tests to see the compatibility between our integrases, including phiBT1 ([[Part:BBa_K2460001|BBa_K2460001]]), Bxb1 ([[Part:BBa_K907000|BBa_K907000]]), phiC31 ([[Part:BBa_K1039012|BBa_K1039012]]), phiRv1 ([[Part:BBa_K2460004|BBa_K2460004]]) and TG1 ([[Part:BBa_K2460007|BBa_K2460007]]) integrase. | ||
| + | |||
| + | ===Genetic design=== | ||
| + | *5 pairs of attB/P sites are each flanking a 50 bp random sequence, lining up ahead of the PBad promoter ([[Part:BBa_I13453|BBa_I13453]]), an RBS ([[Part:BBa_B0033|BBa_B0033]]) and an integrase. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance. | ||
| + | *The same AraC plasmid in Kinetic Characterization was used in orthogonality Characterization. | ||
| + | |||
| + | ===Experimental Setup=== | ||
| + | *The same method of transformation in Kinetic Characterization was used in Kinetic Characterization. | ||
| + | *After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose 16 hours. | ||
| + | *Conduct PCR for each integrase's test and control group with primers from 5 pairs of attB/P sites, 2 pairs of primers for each pair of attB/P site. | ||
| + | |||
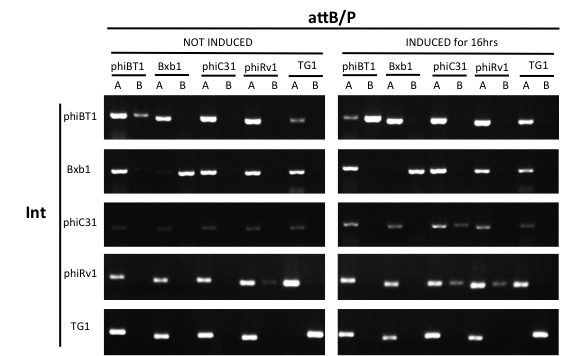
| + | ===Results=== | ||
| + | Expect for the Bxb1 attB/P site, none of the attB/P sites flipped, indicating that Bxb1 integrase has a good orthogonality with other attB/P sites. | ||
| + | |||
| + | [[File:Fudan_China_Orthogonality.jpg]] | ||
| + | |||
| + | |||
| + | [[Part:BBa_K2243000|Click here to get more information about Part:BBa_K2243000!]] | ||
| + | |||
| + | <h2> References:</h2> | ||
| + | <ul> | ||
| + | <li>[1] Salis, Howard M., Ethan A. Mirsky, and Christopher A. Voigt. "Automated design of synthetic ribosome binding sites to control protein expression." Nature biotechnology 27.10 (2009): 946-950. </li> | ||
| + | |||
| + | </ul> | ||
| + | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <span class='h3bb'>Sequence and Features</span> |
| − | <partinfo>BBa_K907000 SequenceAndFeatures</partinfo> | + | <partinfo>BBa_K907000 SequenceAndFeatures</partinfo> |
| − | <!-- Uncomment this to enable Functional Parameter display | + | <!-- Uncomment this to enable Functional Parameter display |
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K907000 parameters</partinfo> | <partinfo>BBa_K907000 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 03:09, 2 November 2017
Mycobacterium Phage Bxb1 gp35, DNA integrase
Characterization by ETH Zurich 2016
- Group: ETH Zurich 2016
- Author: Asli Azizoglu, Lukas Schmidheini
- Summary: We cloned and characterised a codon optimised Bxb1 for E. coli, and sent it to the registry as a biobrick. Our biobrick can be found with a fast degradation tag Part:BBa_K2116009 and here without any degradation tag Part:BBa_K2116026. The non-codon optimised version can be found here Part:BBa_K907000. Besides some favorable features, the codon optimized Bxb1 also supports full assembly compatibility.
Biological Characterisation
Bxb1 is a phage integrase that mediates unidirectional site-specific recombination between two DNA recognition sequences, the phage attachement site, 'attP', and the bacterial attachment site, 'attB'. This recombination event is hereon referred to as a flip. Bxb1 belongs to the family of serine integrases and uses a catalytic serine for strand cleavage, recognizes shorter attP sequences, and does not require host cofactors. It mediates efficient site-specific recombination between two different sequences that are relatively short yet long enough to be specific on a genomic scale. These properties make Bxb1 an efficient tool for creating genetic logic gates, as demonstrated in various papers.
Kinetic Characterisation
We used a reporter construct Part:BBa_K2116024 with a promoter flanked between attB and attP sites Part:BBa_K2116021. Thus, we could investigate the kinetics of Bxb1 using flow cytometry (BD LSR Fortessa SORP) on time course samples.
Genetic Design
- Bxb1 was expressed under a tet promoter, without any degradation tag Part:BBa_K2116056. The RBS has a strength of 1209.69 au (translation initiation rate), as calculated by the Salis Lab RBS Calculator[1]. This construct was cloned on a medium copy plasmid with the p15A replication of origin and Chloramphenicol resistance.
- TetR was expressed under the control of medium-strength constitutive promoter Part:Bba_J23118, and cloned onto a low copy plasmid with pSC101/Rep101 replication of origin and Kanamycine resistance.
- We applied a dual fluorescence reporter system Part:BBa_K2116024 that expresses sfGFP Part:BBa_K2116017 when Bxb1 flips a directional promoter. In the non-flipped state the reporter expresses the red fluorescent protein mNectarine Part:BBa_K2116016.
Experimental Setup
- The test construct with bxb1 (pBR322/rop replication of origin and Ampiciline resistance) was transformed in E. coli TOP10 together with the reporter construct, and either with TetR or with the empty backbone as control.
- TOP10 cells were grown in LB medium (containing Kanamycine, Ampiciline (50 µg/ml) and Chloramphenicol (34 µg/ml)) for 4 hours, and then transfered to minimal M9 medium (containing 0.4% glucose, supplemented with L-leucine, L-isoleucine, and L-valine, Kanamycine, Ampiciline (25 µg/ml and Chloramphenicol (17µg/ml)) with a 1:100 dilution and grown over-night. The culture was again diluted 1:100 in minimal M9 medium the next day. aTc (anhydrotetracycline) was added at given concentrations at OD6000.5. Samples were taken at the specified time points, spun down and resuspended in PBS at OD600 0.01 and kept on ice until measurement.
- Results are presented as mean fluorescence per cell and the standard error of the mean.
Results
We observed that maximum flipping (which we defined as the flipping reached by the used positive controles) is reached at around 11 hours under these conditions. The response correlates with aTc concentration. However, compared to the control, where there is no TetR, we see 6-fold less flipping. This could be improved by increasing the concentration of aTc, however we have found that at 8000 ng/mL cell growth was inhibited.
Comparison between Codon Optimised and Registry Bxb1 Integrase
We compared the kinetics of our codon optimised Bxb1 Part:BBa_K2116026 and the one found previously on the registry Part:BBa_K907000 that was not codon optimized. The same genetic and experimental setup was used as above. The only difference between the two constructs is that the non-codon optimised bxb1 has a weaker RBS, with a strength of 251.82 au (translation initiation rate) [1]. Due to this difference in construction we compared each Bxb1 to its own control (see above, Experimental Setup).
We could show that the codon optimised Bxb1 could reach 85% of maximal flipping, whereas the non-codon optimised one reached only 0.4% after 16 hours of aTc exposure. We thus argue that the codon optimized Bxb1 seems to be more efficient, however, the influence of the different RBS strengths was not properly examined.
Improvement
This part has been improved by iGEM Peking 2017. The improved Part is submitted as BBa_K2243012;https://parts.igem.org/Part:BBa_K2243012
Characterization by Fudan_China 2017
- Group: Fudan_China 2017
- Author: Haiyun Liu, Fudan University
We characterized the kinetic characterization and the orthogonality between Bxb1 and some other serine recombinase. The attB/P site we used for these experiments were Bxb1 attB (BBa_K1039032) and attP (BBa_K1039031)
Kinetic Characterization
We conducted qPCR to record the ratio of attL/R to the sum of attL/R and attB/P, to characterize the efficiency varying with time.
Genetic design
- The integrase is expressed under a PBad promoter (BBa_I13453). Considering the leakage of PBad promoter would lead to flipping,the promoter is followed by a weaker RBS (BBa_B0033). Ahead of the promoter,an attB and attP are opposite to each other,flanking a 50 bp random sequence. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance.
- To reduce leakage when not induced, AraC is expressed under a constitutive promoter, cloned on a high copy plasmid with pUC19 replication of origin and Ampicillin resistance.
Experimental Setup
- Since transforming the two plasmid above at a time was unable to prevent the flipping, the plasmid with AraC was first transformed into E.coli DH10b, then the plasmid with the integrase was transformed into the competent cell with the AraC plasmid.
- After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose for 8 hours. We sampled the culture at 0 hour, the 4th hour and the 8th hour.
- We calculated flipped and unflipped random sequence by qPCR.
Results
Due to the leakage before inducing, the LR rate changing is not obvious, which means the Bxb1 integrase is highly efficient, and the leakage may be reduced by changing the genetic design.
Orthogonality Characterization
Since our circuit are using multiple integrases, it's important that the attB/P site of one integrase won't be flipped by one another integrase. We conducted orthogonality tests to see the compatibility between our integrases, including phiBT1 (BBa_K2460001), Bxb1 (BBa_K907000), phiC31 (BBa_K1039012), phiRv1 (BBa_K2460004) and TG1 (BBa_K2460007) integrase.
Genetic design
- 5 pairs of attB/P sites are each flanking a 50 bp random sequence, lining up ahead of the PBad promoter (BBa_I13453), an RBS (BBa_B0033) and an integrase. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance.
- The same AraC plasmid in Kinetic Characterization was used in orthogonality Characterization.
Experimental Setup
- The same method of transformation in Kinetic Characterization was used in Kinetic Characterization.
- After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose 16 hours.
- Conduct PCR for each integrase's test and control group with primers from 5 pairs of attB/P sites, 2 pairs of primers for each pair of attB/P site.
Results
Expect for the Bxb1 attB/P site, none of the attB/P sites flipped, indicating that Bxb1 integrase has a good orthogonality with other attB/P sites.
Click here to get more information about Part:BBa_K2243000!
References:
- [1] Salis, Howard M., Ethan A. Mirsky, and Christopher A. Voigt. "Automated design of synthetic ribosome binding sites to control protein expression." Nature biotechnology 27.10 (2009): 946-950.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 192
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 466
Illegal XhoI site found at 553 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1105
Illegal NgoMIV site found at 1192
Illegal AgeI site found at 242 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1300