Difference between revisions of "Part:BBa K823030"
| (22 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K823030 short</partinfo> | <partinfo>BBa_K823030 short</partinfo> | ||
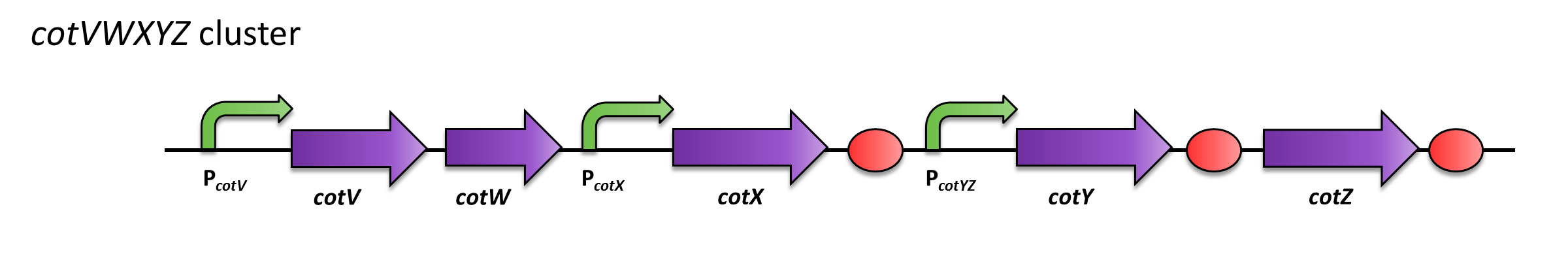
| − | + | The promoter P<sub>''cotYZ''</sub> lies within ''cotVWXYZ'' gene cluster, which regulates expression of CotYZ. The protein CotZ is a componend of the spore crust. Therefore, P<sub>''cotYZ''</sub> is activated in late sporulation. | |
| − | + | ||
| − | + | ||
| − | [[Image:CotVWXYZ cluster.png| | + | ===Usage and Biology=== |
| + | {| style="color:black;" cellpadding="3" width="60%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 60%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[Image:CotVWXYZ cluster.png|Protein distribution in spore coat of ''Bacillus subtilis''|600px|center]] | ||
| + | |- | ||
| + | | style="width: 60%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 60%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2">Gene clusters ''cotVWXYZ''</font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | <br> | ||
| + | <br> | ||
| + | We used this promoter for the expression of spore crust fusion proteins. | ||
| − | |||
| − | + | ===Evaluation=== | |
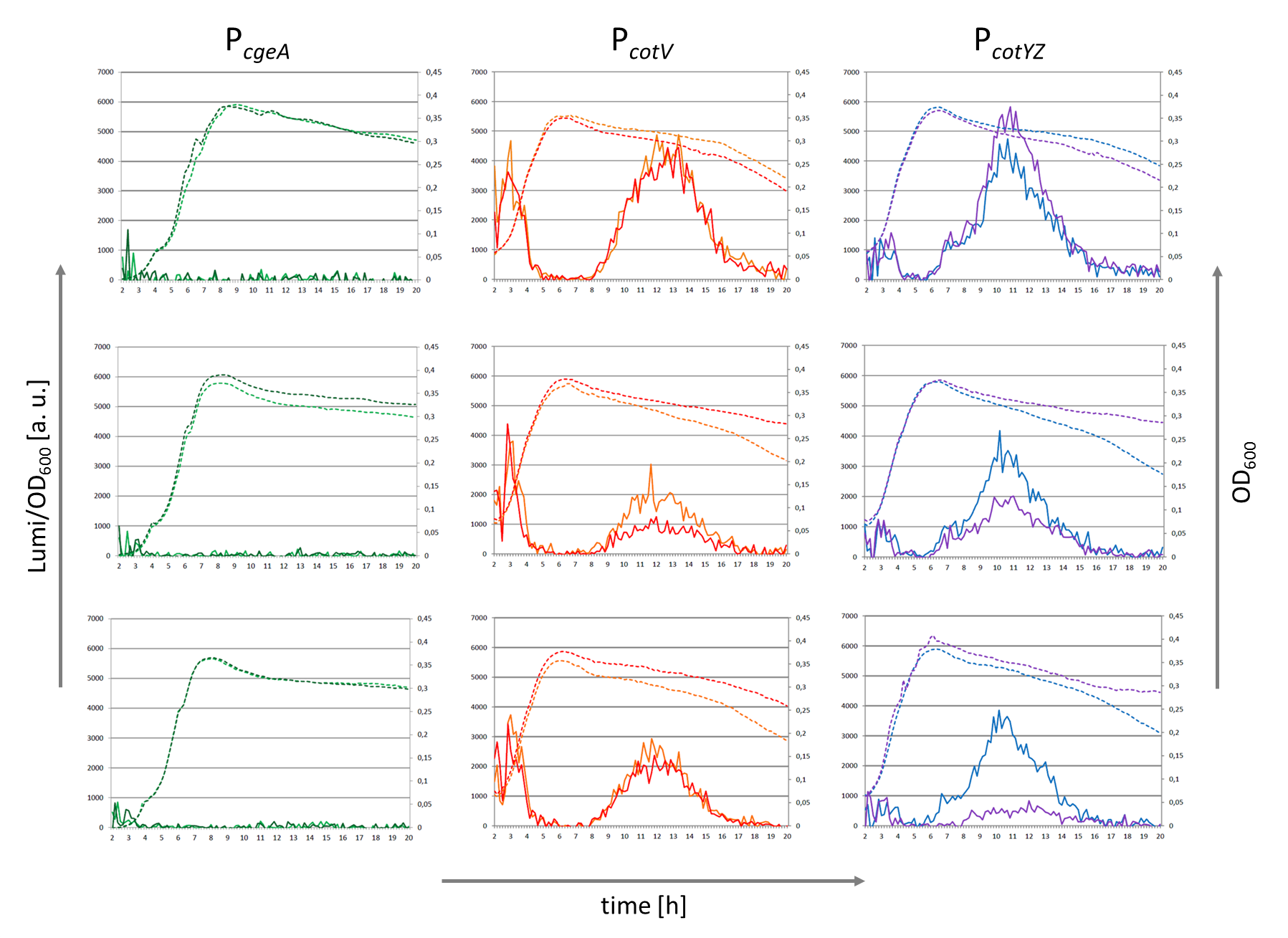
| + | We compared this BioBrick with other promoters that are involved in spore crust formation such as P<sub>''cotV''</sub> and P<sub>''cgeA''</sub>. | ||
| + | To test the activity we cloned this promoter into the lux-reporter vector pSBBS3C-''luxABCDE'' ([[Part:BBa_K823025|BBa_K823025]]) and measured the luminescence. | ||
| − | + | The results of the luminescence measurements are shown in the following figure, which also shows the activities of the related promoters P<sub>''cotV''</sub> ([[Part:BBa_K823033|BBa_K823033]]) and P<sub>''cgeA''</sub>. | |
| − | [[Image:Activity PcotYZ PcotV.png| | + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" |
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[Image:Activity PcotYZ PcotV.png|Protein distribution in spore coat of ''Bacillus subtilis''|610px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="90%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2"><p align="justify">The result of the measurement of this BioBrick P<sub>''cotYZ''</sub> and of two related promoters P<sub>''cgeA''</sub> and P<sub>''cotV''</sub>. Promoters were fused to the lux operon ([[Part:BBa_K823025|pSBBS3C-''luxABCDE'']]) The left vertical axis displays the luminescence divided by the OD<sub>600</sub> and the left one shows the OD<sub>600</sub>. On the horizontal axis the time is illustrated. The continues lines show the luminescence which correlates with the promoter activity and the dotted lines display the growth curve of each clone.</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
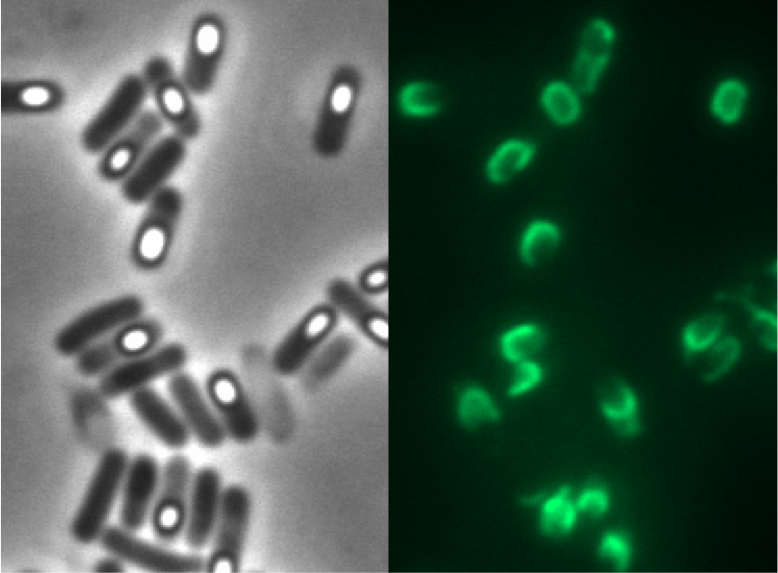
| + | This part was used to display fusion proteins on ''B. subtilis'' spores with [[Part:BBa_K823049|BBa_K823049]]. | ||
| + | [[Image:Glowing spore.png|100px|link=Part:BBa_K823049]] | ||
| − | + | ||
| + | See all the details about the LMU-Munich 2012 project here: | ||
http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins | http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins | ||
| + | |||
| + | [http://2016.igem.org/Team:Freiburg Team Freiburg 2016] improved this part by including a ribosome binding site to the promoter. (see: <partinfo>BBa_K2114000</partinfo>) <br> | ||
| + | (<small>--[[User:WladStr|Wladislaw Stroukov]] 17:32, 12 October 2016 </small>) | ||
Latest revision as of 06:49, 20 October 2016
PcotYZ: B. subtilis promoter regulating expression of spore crust proteins
The promoter PcotYZ lies within cotVWXYZ gene cluster, which regulates expression of CotYZ. The protein CotZ is a componend of the spore crust. Therefore, PcotYZ is activated in late sporulation.
Usage and Biology
|
We used this promoter for the expression of spore crust fusion proteins.
Evaluation
We compared this BioBrick with other promoters that are involved in spore crust formation such as PcotV and PcgeA. To test the activity we cloned this promoter into the lux-reporter vector pSBBS3C-luxABCDE (BBa_K823025) and measured the luminescence.
The results of the luminescence measurements are shown in the following figure, which also shows the activities of the related promoters PcotV (BBa_K823033) and PcgeA.
|
This part was used to display fusion proteins on B. subtilis spores with BBa_K823049.
See all the details about the LMU-Munich 2012 project here: http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins
[http://2016.igem.org/Team:Freiburg Team Freiburg 2016] improved this part by including a ribosome binding site to the promoter. (see: BBa_K2114000)
(--Wladislaw Stroukov 17:32, 12 October 2016 )
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]