Difference between revisions of "Part:BBa K729002"
| (70 intermediate revisions by 12 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K729002 short</partinfo> | <partinfo>BBa_K729002 short</partinfo> | ||
| − | + | Laccase for the degradation of polyethylene and organic pollutants. Sequence pending. | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 16: | ||
<partinfo>BBa_K729002 parameters</partinfo> | <partinfo>BBa_K729002 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Description=== | ||
| + | The laccase enzyme has been shown to degrade polyethylene when being expressed by a number of different bacteria and fungi. This is due to conserved copper binding sites which couple the oxidation of a substrate with the cleavage of dioxygen bonds, leading to the capability to degrade plastics (particularly polyethylene). | ||
| + | |||
| + | By driving the laccase production using a strong constitutive promoter, we expect overexpression of the protein which will allow it to be secreted into the extracellular medium where it can act upon the target plastic. | ||
| + | |||
| + | ===Characterisation=== | ||
| + | |||
| + | Plastic degradation is mediated via a laccase protein. As such, we will be using an enzymatic activity assay to determine that the laccase enzyme is expressed. Laccase catalyses the oxidation of <span class="footnote" title="Syringaldazine">syringaldazine</span>, a reaction that exhibits an observable OD change at 530nm. A sample of syringaldazine can be used as a blank in a spectrophotometer, against a sample containing syringaldazine and our laccase sample, allowing the rate of oxidation to be measured, and hence the enzymatic activity of laccase. | ||
| + | |||
| + | In order to determine the effectiveness of laccase in degrading plastic, we will expose strips of various types of plastic to the laccase expressing bacteria, before viewing the strips under a scanning electron microscope. This will allow us to compare the pitting in plastic samples treated with laccase, to the untreated samples, allowing us to determine the extent of plastic degradation. | ||
| + | |||
| + | ===Results=== | ||
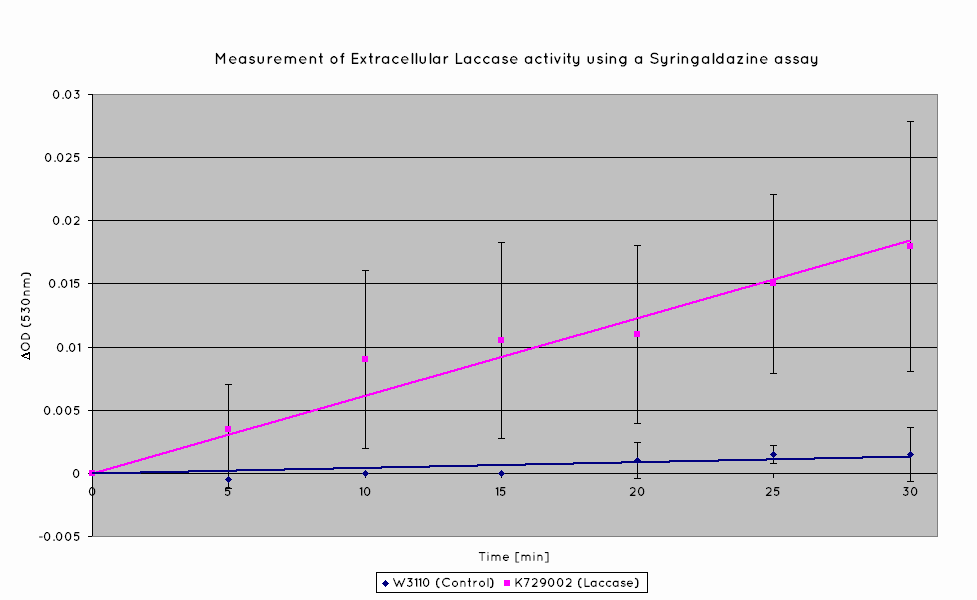
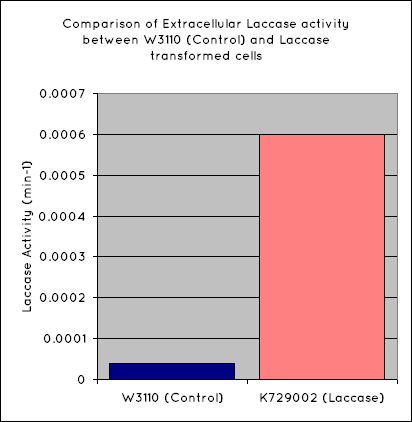
| + | Our results indicate a significantly higher rate of oxidation from the cells containing our BioBrick than the control cell line. This indicates that our transformed ''E. Coli'' have successfully produced laccase, and in significant enough quantities that it is released into the extracellular space. This allows it to oxidise the syringaldazine utilised in the laccase assay. | ||
| + | |||
| + | [[Image:UniversityCollegeLondon_Laccase_Assay_Extracellular.png|600px]] | ||
| + | [[Image:UniversityCollegeLondon_Laccase_Activity.png|300px]] | ||
| + | |||
| + | |||
| + | The Scanning Electron Microscope (SEM) images below show the effects of subjecting Low Density Polyethylene (LDPE) to three different sets of conditions in order to visualise the degradative properties of cells containing our Laccase construct (BBa_K729006). We can then also compare this with the ability of untransformed ''E. coli'' W3110 and mechanical shear in water to breakdown the LDPE target. | ||
| + | |||
| + | All samples were in suspension at 37°C and 200rpm in an incubated shaker for 3 days. | ||
| + | |||
| + | Low magnification (x100) images were used to observe the macro-trend in zonal enzymatic activity. There appears to be increased ‘roughness’ (shown by the small protrusions on the surface as well as areas of darker shading, which represent areas of greater contour), across the surface of the plastic as we progress from the sample treated with water to untransformed ''E. coli'' W3110 to ''E. coli'' transformed with the UCL Laccase construct. | ||
| + | |||
| + | High magnification (x10 000) images were taken to give more high resolution information regarding the activity of the enzyme on a much smaller scale. The SEM images highlight increased amounts of ‘pitting’ (the generation of small holes in the plastic due to it’s structural breakdown), as we go from water to ''E. coli'' W3110 to bacteria containing BBa_K729006. These pits are shown by the much darker areas. This small scale breakdown has a cumulative effect which results in the overall trend seen at the lower magnification. | ||
| + | |||
| + | In conclusion, this collection of SEM images indicates an ability both of the BBa_K729006 transformed cells to produce the Laccase enzyme at greater titres than untransformed bacteria (though the ''E. coli'' W3110 is still seen to have some degradative effect), and of the Laccase produced to degrade the polyethylene target. | ||
| + | |||
| + | |||
| + | {| border="1" | ||
| + | |+ '''SEM Results''' | ||
| + | ! !! Water !! W3110 !! Laccase | ||
| + | |- | ||
| + | ! Low Magnification (100x) | ||
| + | | [[Image:UniversityCollegeLondon_LaccaseSEM_Water_Low_mag.png|280px]] || [[Image:UniversityCollegeLondon_LaccaseSEM_W3110_Low_mag.png|280px]] || [[Image:UniversityCollegeLondon_LaccaseSEM_Laccase_Low_mag.png|280px]] | ||
| + | |- | ||
| + | ! High Magnification (10 000x) | ||
| + | | [[Image:UniversityCollegeLondon_LaccaseSEM_Water_High_mag.png|280px]] || [[Image:UniversityCollegeLondon_LaccaseSEM_W3110_High_mag.png|280px]] || [[Image:UniversityCollegeLondon_LaccaseSEM_Laccase_High_mag.png|280px]] | ||
| + | |} | ||
| + | |||
| + | ===Modelling=== | ||
| + | |||
| + | ===Conclusion=== | ||
| + | |||
| + | Through the use of a simple quantitative assay, it has been ascertained that ''E. coli'', transformed with our construct has a significantly increased extracellular laccase activity when compared with a control cell line. Combined with the results from the scanning electron microscope, which indicate surface degradation of polyethylene in a relatively short timespan, indicates that this BioBrick holds powerful potential for the break down of polyethylene. | ||
| + | |||
| + | =iGEM MITADTBIO_Pune 2019 characterization= | ||
| + | |||
| + | |||
| + | ===Description=== | ||
| + | <i>Laccase</i> enzyme acts on many different substrates, with its activity also prominent in helping break down polyethylene. Since our aim is to degrade plastic present in menstrual sanitary napkins, it was important to know whether it's activity is affected in the presence of blood. Laccase has mutiple copper ions in its cofactor and therefore it was crucial for us to know whether the activity of laccase is hindered in the presence of other ions present in blood. | ||
| + | |||
| + | ===Cloning of laccase gene from BBa K729002 into pET28a+=== | ||
| + | |||
| + | Cloning of <i>laccase</i> in iGEM backbone plasmid will lead to normal low level of <i>laccase</i> expression which will show <i>in vitro</i> activity, but the activity and expressive rate is non-obvious. For the above reasons, we decided to clone laccase gene is pET28a+ expression vector for sustainable production of <i>laccase</i>. <br> | ||
| + | To insert <i>laccase</i> gene into cloning expression vector (pET 28a+), we first designed prefix and suffix primers which contain <i>EcoRI</i> and <i>PstI</i> restriction enzyme sites respectively. The PCR amplification of <i>laccase</i> gene from BBa_K729002 using <i>EcoRI</i>-prefix and <i>PstI</i>-suffix primers give an expected 1.5 kb DNA band as shown in (Fig. 1). | ||
| + | <br> | ||
| + | <center>https://2019.igem.org/wiki/images/f/fd/T--MITADTBIO_Pune--ORF.png</center> | ||
| + | <br> | ||
| + | <center><b>Figure 1. PCR amplification of laccase ORF using prefix and suffix primers gave expected size DNA band. M: 500 bp DNA ladder; 1: PCR amplified laccase gene (size 1.5 kb).</b></center> | ||
| + | <br> | ||
| + | The purified PCR amplified products were restriction double digested with <i>EcoRI-PstI</i>, and ligated at same sites in pET28a+ (5.3 kb) expression vector and transformed recombinant plasmid into <i>E. coli</i> (JM109). The transformation result was shown in Figure 2. To check whether the construction was successful, we picked few colonies to extract their plasmids. The inclusion of insert into the recombinant plasmid was verified both by restriction digestion. | ||
| + | <center>https://2019.igem.org/wiki/images/4/46/T--MITADTBIO_Pune--pet1.png</center> | ||
| + | <center><b>Figure 2. Restriction digestion of <i>laccase</i> carrying recombinant plasmid using <i>EcoRI</i>. M: 500 bp DNA ladder; 1: Plasmid showing expected size band of 1.55 kb.</b></center> | ||
| + | <br> | ||
| + | We tested the expression of <i>laccase</i> enzyme by transforming the pET28a+ recombinant carrying pET-laccase gene into <i>E. coli</i> expression strain BL21. To overexpress <i>laccase</i> enzyme, <i>E. coli</i> expression strain BL21 containing <i>laccase</i> gene under IPTG-inducible T7 promoter was induced by 1mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3hrs and 18 hrs at 37°C, corresponding proteins were separated by 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue G250 (CBB). Prior to induction with IPTG, expression of <i>laccase</i> is not detectable by SDS-PAGE (Figure 3 Lane 1). After IPTG induction recombinant, protein bands of about 58 kDa was observed which is in agreement with the theoretical molecular weight of laccase which is 58.87 kDa (Figure 3 Lane 2). | ||
| + | <center>https://2019.igem.org/wiki/images/0/02/T--MITADTBIO_Pune--SDS.png</center> | ||
| + | <br> | ||
| + | <center><b>Figure 3. Induction and overexpression of recombinant protein: Samples were subjected to 12% SDS-PAGE and stained with Commassie Brilliant Blue G250. A- Lane 1, total cellular proteins from <i>E. coli</i> BL21(DE3) transformed with pET-laccase uninduced; Lane 2, total cellular proteins from <i>E. coli</i> BL21(DE3) transformed with pET-laccase induced by IPTG for 3 hrs; Lane 2, total cellular proteins from <i>E. coli</i> BL21(DE3) transformed with pET-laccase induced by IPTG for 18 hrs.</b></center> | ||
| + | <br> | ||
| + | ===<i>In vitro</i> laccase assay=== | ||
| + | We tested whether expressed laccase is active through enzyme activity experiments. To detect laccase enzyme activity, colorimetric assay was carried out using 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) or ABTS method. | ||
| + | The nonphenolic dye ABTS is oxidized by laccase into water soluble chromogen ABTS+ which is more stable and preferred state of the cation radical. ABTS+ can be monitored spectrophotometrically at 420 nm. The concentration of the cation radical responsible for the intense blue-green color can be correlated to enzyme activity and is read at 420nm. The supernatant of overnight grown bacteria culture acted as a source of laccase enzyme. The enzyme activity experiment was carried out by using 100 μL of enzyme and 100 μL of ABTS solution (0. 5 mM) in sodium acetate buffer (pH 5). The enzymatic units (U) defined as the amount of enzyme transforming 1 µmol of substrate per minute is calculated by the following formula: | ||
| + | Equation : U/L = (∆E×Vt)/(ε×d×Vs) | ||
| + | |||
| + | with ∆E being the change in the extinction of light [min-1] at 420 nm<br> | ||
| + | ε being the molar absorption coefficient of ABTS [M-1 cm-1]<br> | ||
| + | d being the layer thickness [cm] in your cell that the light has to pass<br> | ||
| + | Vt is the total volume measured and Vs is the volume of the enzyme stock solution added to the ABTS stock solution.<br> | ||
| + | It is demonstrated in the given graph by in vitro ABTS assay that recombinant laccase protein can oxidize ABTS. It is essential for our project that recombinant microorganisms should be capable of polyethylene deterioration in the presence of blood and/or dried blood. To check the effect of blood on laccase enzyme activity, we performed the ABTS assay in the presence of different concentrations of blood. | ||
| + | |||
| + | <center>https://2019.igem.org/wiki/images/d/d0/T--MITADTBIO_Pune--plot.png</center> | ||
| + | <b><center>Figure 4. Laccase activity in presence and absence of blood. The fold differences were calculated compared with control groups and shown in the figure. *P<0.007; **P<0.01; [one-way analysis of variance].</center></b> | ||
| + | |||
| + | ===Conclusion=== | ||
| + | It was clearly observed that the laccase activity is not significantly affected by blood. | ||
| + | |||
| + | ==Added by LZU-HS-China-C== | ||
| + | |||
| + | We found that the laccase BBa_K729002 may also have the effect of degrading antibiotics. So we used this laccase to complete the verification experiment of degrading antibiotics. | ||
| + | |||
| + | At the same time, in order to enhance its function, we added INP protein to the N segment of BBa_K729002, so that the laccase was anchored on the surface of the bacteria through cell surface display technology. | ||
| + | |||
| + | In order to facilitate the comparison of the degradation ability of BBa_K729002 and BBa_K4332002 with INP added, we will compare and show these two parts. | ||
| + | |||
| + | (Correspondence between the gene in the picture and the gene in part:BBa_K729002----lacc6、BBa_K4332002----IL) | ||
| + | |||
| + | ===Usage and Biology=== | ||
| + | Laccase is an oxidoreductase that uses oxygen as an electron acceptor. It can oxidize a variety of aromatic compounds and produce water as a by-product. In the 19th century, humans first discovered laccase in Japanese sumac. Because of its outstanding application value in biotechnology applications, scientists have gradually deepened their research on laccase. They discovered that laccase is also present in animals and microorganisms. Laccases derived from different hosts have different functions in the body. Among them, fungal laccases derived from white rot fungi, Trametes, Pleurotus ostreatus and other fungal laccases occupy the mainstream of the current laccase application process. This type of laccase has a relatively wide range of substrate specificity during the application process, and does not require hydrogen peroxide. With the participation of, a variety of phenolic compounds and their derivatives can be directly oxidized with oxygen as the electron acceptor. | ||
| + | |||
| + | In recent years, the main methods for removing antibiotic residues from different sources are traditional methods such as high-temperature composting, fermentation; oxidation, adsorption, electrochemical treatment, membrane method, and microbial degradation. However, these methods have the disadvantages of low removal efficiency, high cost of use, and the possibility of secondary pollution to the environment. Moreover, these methods cannot fundamentally solve the problem of antibiotic residues in poultry. Therefore, it is necessary to find a direct, effective and low-cost method to remove antibiotic residues in poultry. At present, many studies have shown that fungal laccase plays an important role in the process of treating antibiotic residues in wastewater. Three laccase-producing genes LACC6, LACC9, and LACC10 obtained from Pleurotus ostreatus were heterologously expressed in Pichia pastoris and found that the efficiency of degrading sulfa antibiotics reached more than 97%. | ||
| + | |||
| + | ===Heterologous expression and application advantages of fungal laccase=== | ||
| + | Studies have shown that fungal laccase is a multi-copper oxidase that can deliver 4 copper ions at the same time, so it can catalyze a variety of phenolic compounds and aromatic compounds. Based on its wide range of substrates, laccase has become a useful biocatalyst in the application of biotechnology, especially in the field of bioremediation, including the degradation of lignin, the decolorization of synthetic dye wastewater, and the degradation and detoxification of environmental pollutants. However, the laccase secreted by wild strains is limited in yield and difficult to purify, on the other hand, its activity is susceptible to environmental factors. In recent years, microbial fermentation engineering has gradually emerged, and the fermentation production of most laccases mainly stays at the level of shake flasks and fermentation tanks. Ryan et al. used an air-lift fermentor to ferment Trametes villus, and the laccase activity in the fermentation broth could reach 11.8 U/ml. At this stage, although the production and application of some fungal laccases can be realized by using the fermentation process, the culture conditions of microorganisms such as temperature, pH, humidity, and the length of the fermentation cycle will affect the production of laccase during the fermentation process. Therefore, most laccases are fermented The process only stays at the laboratory scale. Based on the above shortcomings, it is very important to develop an efficient, low-cost, and environmentally friendly method for producing laccase. | ||
| + | |||
| + | At this stage, researchers have used molecular biology methods to heterologously express laccase genes on the basis of overcoming many shortcomings of traditional laccase purification processes and fermentation processes, in order to obtain laccase with higher yield and enzymatic activity. At present, according to production requirements, laccases from different sources have been successfully expressed in various host cells such as bacteria, fungi, and insect rod cells. At present, in industrial applications, fungal cells such as Pichia pastoris, Saccharomyces cerevisiae and Kluyveromyces lactis dominate the heterologous expression process of laccase. These fungal cells have the following advantages in the application process: first, high-density fermentation can increase the production of laccase, second, the engineered strain has higher enzyme activity, and third, most fungal host cells have been confirmed to be directly edible by humans. | ||
| + | |||
| + | [[Image:T--LZU-HS-China-C--China-C4.jpg | thumb | center | 800px |Figure 1 ]] | ||
| + | |||
| + | In recent years, with the deepening of research, scientists have found that laccase also has many shortcomings in fungal heterologous expression. When the engineered strain is fermented, it is found that the fermentation cycle is longer and usually takes about one week, and the strain culture consumes a lot. In their research, Maestre et al. found that host yeast can inactivate laccase by interfering with the glycosylation metabolism of laccase. Laccase-producing bacteria from the same source often have multiple laccase genes at the same time, and the existence of these genes ultimately leads to a more complex and diverse isozyme system of laccase. Therefore, when the researchers performed heterologous expression of these isoenzymes, they found that not all fungal expression systems can maximize the productivity of laccase, and the laccase activity produced by heterologous expression in yeast cells is far less than that of wild-type laccase. Strains are also susceptible to factors such as pH. Compared with wild strains, the half-life of laccase in some expression systems has also changed. Comprehensive analysis of the heterologous expression of laccase in fungi, the bacterial expression system has been gradually applied to the direction of heterologous expression of laccase due to its easy operation, short culture period, and strong reproductive ability. | ||
| + | |||
| + | ===The ability of fungal laccase to degrade sulfadiazine=== | ||
| + | EcN-Lacc6 refers to the fungal laccase gene expressed in the cell | ||
| + | |||
| + | This experiment mainly uses HPLC to quantitatively analyze the residues of sulfadiazine. The process involves two parts: sample pretreatment and HPLC on-machine detection. The specific experimental method is as follows; | ||
| + | |||
| + | (1) Sample pretreatment. First, EcN-IL and the control wild-type EcN, the unloaded EcN and the intracellular expression strain EcN-Lacc6 were inoculated into LB medium at 37°C overnight at an inoculum of 1%. After centrifugation to remove the supernatant, the bacteria were collected (6000 rpm, 10min). The bacteria were washed twice with the same amount of PBS (pH=7.0) buffer solution and prepared into a bacterial suspension with a cell concentration of 1.0 × 109 CFU/mL. Take 10 mL of the cell suspension and add SDZ sodium salt with a concentration of 30, 50, and 100 mg/L into it, with three replicates in each group. Incubate at room temperature for 3 hours and then centrifuge (10000 r/min, 10 min), collect the supernatant into a 10mL PE tube, and finally filter it into a special brown sample bottle for HPLC using a 0.22μm filter membrane. | ||
| + | |||
| + | (2) HPLC on-machine detection conditions selection. HPLC loading conditions selection: select VWD detector, detection wavelength is set to 270 nm, methanol and ultrapure water are organic phase, mobile phase: methanol: water=25:75, detection column Waters XTERRA MS C18 column (250× 4.6 mm), the column temperature is set to 30°C, and the mobile phase flow rate is 1.0 mL/min. HPLC detection data is calculated using external standard method. | ||
| + | |||
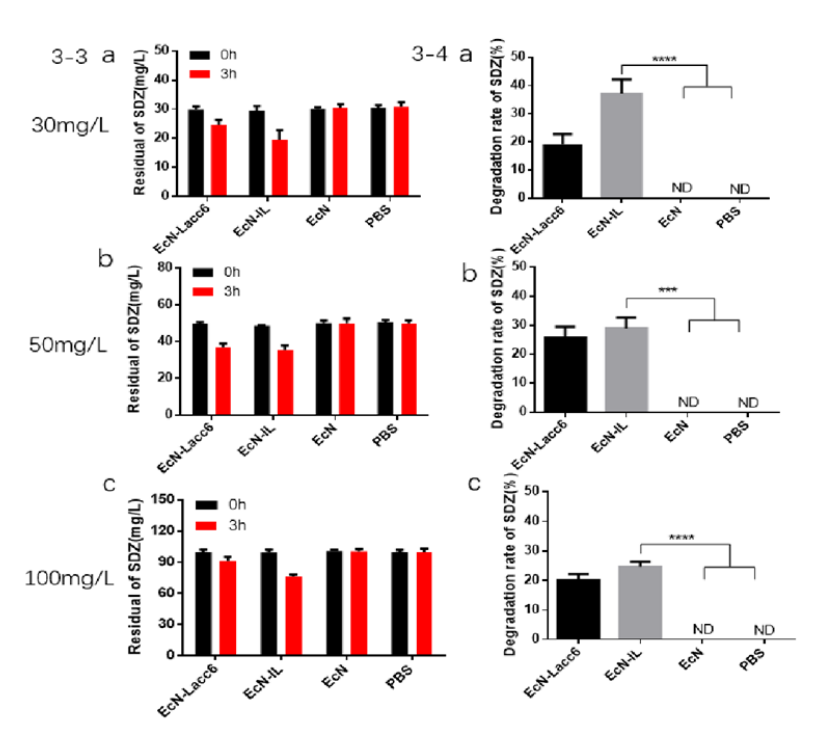
| + | [[Image:T--LZU-HS-China-C--China-C5.jpg | thumb | center | 500px |Figure 2 ]] | ||
| + | |||
| + | a, b, c show the degradation rate analysis of EcN-IL to sulfadiazine at different concentrations (30, 50 and 100 mg/L). Compared with the control EcN group, the degradation rate of EcN-IL to SDZ can reach up to 30 ±2.3%. | ||
| + | |||
| + | ==Contribution: Hong Kong JSS 2021== | ||
| + | ===Usage and Biology=== | ||
| + | In iGEM competition, 2012 Team UCL London, 2012 Team Bielefeld-Germany, 2018 Team HKUST, and 2019 Team MITADTBIO have explored the use of laccase to degrade Polyethylene plastic <html>(<a href="https://parts.igem.org/Part:BBa_K729002">BBa_K729002</a>)</html> and showing the enzymatic property of laccase <html>(<a href="https://parts.igem.org/Part:BBa_K863005">BBa_K863005</a>)</html>. The teams have demonstrated that the laccase protein can be heterogeneously expressed in <i>E. coli</i> and being secreted to the extracellular spaces. | ||
| + | |||
| + | In our project, we focused on the aflatoxin B1 degrading ability of laccase. From our knowledge, there is no iGEM project addressing the AFB1 degradation ability of laccase yet. | ||
| + | |||
| + | Aflatoxin (AF) is a family of carcinogenic toxins produced by <i>Aspergillus</i> sp.. According to the World Health Organization (WHO), 25% of food crops are destroyed due to aflatoxin contamination each year. About 5 billion people are at risk of chronic AF exposure and more than 80% of them will develop AF-related diseases such as hepatocellular carcinoma and liver failure. [2] Among all AF, aflatoxin B1 (AFB1) is considered the most potent and chronic. [3] | ||
| + | |||
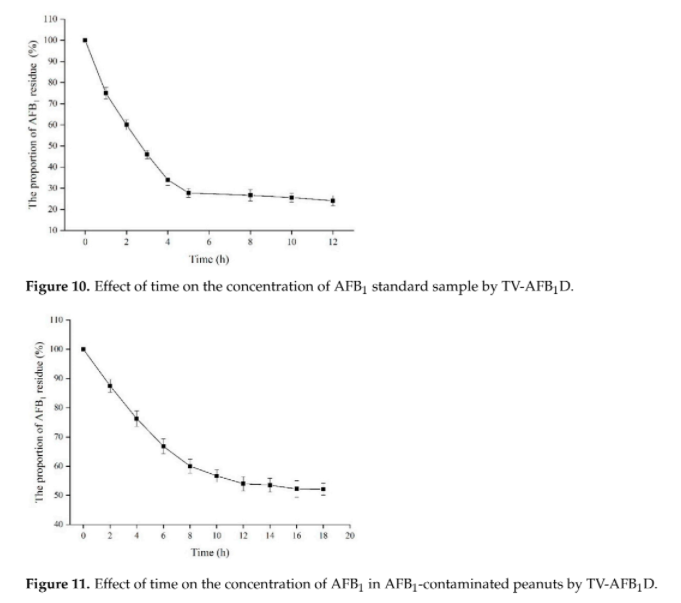
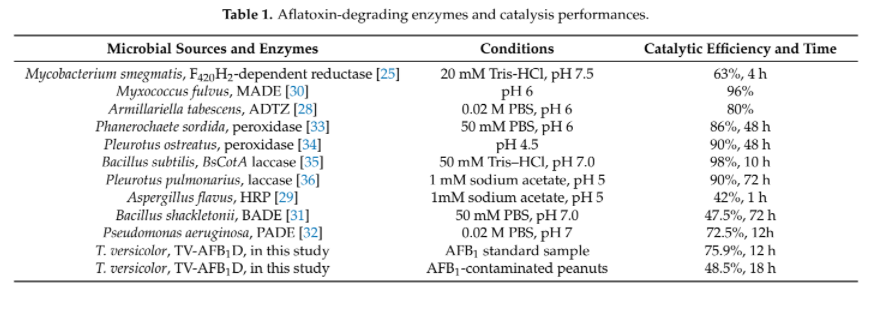
| + | Different groups have demonstrated the effectiveness of AFB1 degradation by using native and recombinant laccase [4]. While Yang, P. et al. 's group in 2021 has demonstrated that recombinant laccase produced from <i>P. pastoris</i> could decrease ~48.5% of AFB1 in peanut samples after 18 hour. (Figure 1)[7] This result further support our plan that a detoxifying spray of laccase can help to tackle the problem of AFB1 contaminated food. | ||
| + | |||
| + | [[File:TvLac AFB1 efficiency.png|500px|thumb|center|Fig. 1 The heterogeneously expressed recombinant tvLac can reduce the concentration of AFB1 in pure AFB1 substrate and peanut substrates effectively. Table adopted from Yang, P., et al. (2021)[7]]] | ||
| + | |||
| + | Although there are other enzymes that were found to have AFB1 degrading ability, but they either shown lower efficacy or lack of sufficient studies to prove their function when they are heterogeneously expressed. (Table 2). Thus, laccase is one of the most well-studied target that aroused our interest to study in our project. | ||
| + | |||
| + | [[File:Compare AFB1-enzyme activity.png|500px|thumb|center|Fig. 2 The comparison of different enzyme's activity in AFB1 degradation. Table adopted from Yang, P., et al. (2021)[7]]] | ||
| + | |||
| + | Among all the laccase produced by different species reviewed, laccase produced by <i>Trametes versicolor</i> (white rot) was found to be the most effective in AFB1 degradation. [3,5] Thus, our team proposes to heterogeneously express this tvLac in probiotic <i>E. coli</i> and to see if it can be a plausible measure to degrade AFB1 in food and act as a food preservative for <i>Aspergillus</I> sp. infection. | ||
| + | |||
| + | We have decided a basic part coding for tvLac for our 2021 phase 1 iGEM project. <html>(<a href="https://parts.igem.org/Part:BBa_K3746002">BBa_K3746002</a>)</html> and expression composite parts of FDR-A <html>(<a href="https://parts.igem.org/Part:BBa_K3746006">BBa_K3746006</a>)</html>, <html>(<a href="https://parts.igem.org/Part:BBa_K3746007">BBa_K3746007</a>)</html>, and <html>(<a href="https://parts.igem.org/Part:BBa_K3746009">BBa_K3746009</a>)</html>. | ||
| + | |||
| + | ===Reference=== | ||
| + | [1] Sumathi T, Viswanath B, Sri Lakshmi A, SaiGopal DV. Production of Laccase by Cochliobolus sp. Isolated from Plastic Dumped Soils and Their Ability to Degrade Low Molecular Weight PVC. Biochem Res Int. 2016;2016:9519527. doi: 10.1155/2016/9519527. Epub 2016 May 12. PMID: 27293894; PMCID: PMC4880699. | ||
| + | |||
| + | [2] Organization WH. aflatoxin. Manuf Comput Solut. 2000;6(8):20-3 | ||
| + | |||
| + | [3] Okwara, P. C., Afolabi, I. S., & Ahuekwe, E. F. (2021). Application of laccase in aflatoxin B1 degradation: A Review. IOP Conference Series: Materials Science and Engineering, 1107(1), 012178. https://doi.org/10.1088/1757-899x/1107/1/012178 | ||
| + | |||
| + | [4] Alberts, J. F., Gelderblom, W. C. A., Botha, A., & van Zyl, W. H. (2009). Degradation of aflatoxin B1 by fungal laccase enzymes. International Journal of Food Microbiology, 135(1), 47–52. https://doi.org/10.1016/j.ijfoodmicro.2009.07.022 | ||
| + | |||
| + | [5] Verheecke, C., Liboz, T., & Mathieu, F. (2016). Microbial degradation of aflatoxin B1: Current status and future advances. International Journal of Food Microbiology, 237, 1–9. https://doi.org/10.1016/j.ijfoodmicro.2016.07.028 | ||
| + | |||
| + | [6] Guo, Y., Qin, X., Tang, Y., Ma, Q., Zhang, J., & Zhao, L. (2020). Cota Laccase, a novel aflatoxin oxidase from bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chemistry, 325, 126877. https://doi.org/10.1016/j.foodchem.2020.126877 | ||
| + | |||
| + | [7] Yang, P., Xiao, W., Lu, S., Jiang, S., Zheng, Z., Zhang, D., Zhang, M., Jiang, S., & Jiang, S. (2021). Recombinant expression of trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) in engineering pichia Pastoris GS115 and application in AFB1 degradation in AFB1-contaminated peanuts. Toxins, 13(5), 349. https://doi.org/10.3390/toxins13050349 | ||
| + | |||
| + | |||
| + | ==Team TecCEM 2022 detailed enzyme description== | ||
| + | <html> | ||
| + | <br> | ||
| + | <strong><font size=5>Laccase of <em>Trametes</em> Versicolor </font></strong> | ||
| + | <p>iGEM Tec CEM 2022’s construct</p> | ||
| + | <p></p> | ||
| + | <H3>Summary</H3> | ||
| + | <p align = "justify">The amino acid sequence was entered in the iTASSER platform to know the structures. Thanks to this model and the subsequent visualization in PYMOL, it was possible to identify the amino acids that are part of the active site, the secondary structures that give it form, and the possible formation of disulfide bonds. Likewise, this model was used to evaluate the pHs at which laccase is most stable, and finally the isoelectric point, to determine its charge at different pHs.</p> | ||
| + | |||
| + | <center> | ||
| + | <img style="vertical-align: bottom;)" width=50% src="https://static.igem.org/mediawiki/parts/f/fa/K729002_TecCem_Fig1.png"> | ||
| + | </center> | ||
| + | <center><strong>Fig. 1 </strong>Crystallized image of iGEM 2022 laccase. Secondary structures and protein surface are observed.</center></strong> | ||
| + | <br><strong>Number of amino acids</strong></br> | ||
| + | <br>535</br> | ||
| + | <br><strong>Molecular Weight</strong></br> | ||
| + | <br>58521.33 Da</br> | ||
| + | |||
| + | <br><strong>Isoelectric Point</strong></br> | ||
| + | <br>(pI)6.44</br> | ||
| + | |||
| + | <br><strong>Extinction Coefficient</strong></br> | ||
| + | <table> | ||
| + | <tr></tr> | ||
| + | <tr> | ||
| + | <td>Cys fully reduced</td> | ||
| + | <td></td> | ||
| + | <td>62910.00 M<sup>-1</sup>cm<sup>-1</sup></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Abs 0.1% (1 g/l)</td> | ||
| + | <td></td> | ||
| + | <td>1.075</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Cys fully oxidized</td> | ||
| + | <td></td> | ||
| + | <td>63035.00 M<sup>-1</sup>cm<sup>-1</sup></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Abs 0.1% (1 g/l) </td> | ||
| + | <td></td> | ||
| + | <td>1.077</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br><strong>Instability Index</strong></br> | ||
| + | <br>31.54 (stable)</br> | ||
| + | <br><strong>Optimal pH</strong></br> | ||
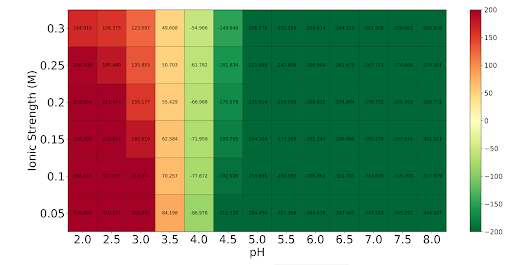
| + | <p align = "justify">The PDB file of the modeled laccase was obtained in iTASSER and entered into the Heatmap tool of Protein-sol. Based on the heatmap, its optimal pH is between 5.0 and 8.0, according to the green coloration.</p> | ||
| + | <center> | ||
| + | <img style="vertical-align: bottom;)" width=45% src="https://static.igem.org/mediawiki/parts/b/bc/K729002_TecCem_Fig2.png"> | ||
| + | </center> | ||
| + | <center><strong>Fig. 2</strong> Energy heatmap ( J per aa).</center> | ||
| + | |||
| + | <br><strong>Isoelectric Point</strong></br> | ||
| + | <p align = "justify">Based on the heatmap, its PI is close to pH 6.9, due to the white coloration and almost zero charges. Both heatmaps were obtained from <a href="https://protein-sol.manchester.ac.uk/heatmap">https://protein-sol.manchester.ac.uk/heatmap</a></p> | ||
| + | <center> | ||
| + | <img style="vertical-align: bottom;)" width=45% src="https://static.igem.org/mediawiki/parts/0/0a/K729002_TecCem_Fig3.png"> | ||
| + | </center> | ||
| + | <center><strong>Fig. 3</strong> Charge heatmap (e per aa).</center> | ||
| + | <br><strong>Amino Acids frequencies</strong></br> | ||
| + | <br> | ||
| + | <table> | ||
| + | <tr><td>Ala</td><td></td><td>A</td><td></td><td>50</td><td></td><td>9.3%</td></tr> | ||
| + | <tr><td>Arg</td><td></td><td>R</td><td></td><td>20</td><td></td><td>3.7%</td></tr> | ||
| + | <tr><td>Asn</td><td></td><td>N</td><td></td><td>21</td><td></td><td>3.9%</td></tr> | ||
| + | <tr><td>Asp</td><td></td><td>D</td><td></td><td>31</td><td></td><td>5.8%</td></tr> | ||
| + | <tr><td>Cys</td><td></td><td>C</td><td></td><td>3</td><td></td><td>0.6%</td></tr> | ||
| + | <tr><td>Gln</td><td></td><td>Q</td><td></td><td>24</td><td></td><td>4.5%</td></tr> | ||
| + | <tr><td>Glu</td><td></td><td>E</td><td></td><td>21</td><td></td><td>3.9%</td></tr> | ||
| + | <tr><td>Gly</td><td></td><td>G</td><td></td><td>52</td><td></td><td>9.7%</td></tr> | ||
| + | <tr><td>His</td><td></td><td>H</td><td></td><td>23</td><td></td><td>4.3%</td></tr> | ||
| + | <tr><td>Ile</td><td></td><td>I</td><td></td><td>22</td><td></td><td>4.1%</td></tr> | ||
| + | <tr><td>Leu</td><td></td><td>L</td><td></td><td>56</td><td></td><td>10.4%</td></tr> | ||
| + | <tr><td>Lys</td><td></td><td>K</td><td></td><td>26</td><td></td><td>4.9%</td></tr> | ||
| + | <tr><td>Met</td><td></td><td>M</td><td></td><td>31</td><td></td><td>5.8%</td></tr> | ||
| + | <tr><td>Phe</td><td></td><td>F</td><td></td><td>18</td><td></td><td>3.4%</td></tr> | ||
| + | <tr><td>Pro</td><td></td><td>P</td><td></td><td>35</td><td></td><td>6.5%</td></tr> | ||
| + | <tr><td>Ser</td><td></td><td>S</td><td></td><td>21</td><td></td><td>3.9%</td></tr> | ||
| + | <tr><td>Thr</td><td></td><td>T</td><td></td><td>27</td><td></td><td>5.0%</td></tr> | ||
| + | <tr><td>Trp</td><td></td><td>W</td><td></td><td>9</td><td></td><td>1.7%</td></tr> | ||
| + | <tr><td>Tyr</td><td></td><td>Y</td><td></td><td>9</td><td></td><td>1.7%</td></tr> | ||
| + | <tr><td>Val</td><td></td><td>V</td><td></td><td>37</td><td></td><td>6.9%</td></tr> | ||
| + | <tr><td>Pyl</td><td></td><td>O</td><td></td><td>0</td><td></td><td>0.0%</td></tr> | ||
| + | <tr><td>Pyl</td><td></td><td>U</td><td></td><td>0</td><td></td><td>0.0%</td></tr> | ||
| + | </table> | ||
| + | </br> | ||
| + | |||
| + | <br><strong>Amino acid sequence:</strong></br> | ||
| + | <p align ="justify">MKKITAAAGLLLLAAQPAMAMQRRDFLKYSVALGVASALPLWSRAVFAAERPTLPIPDLLTTDARNRIQLTIGAGQSTFGGKTATTWGYNGNLLGPAVKLQRGKAVTVDIY NQLTEETTLHWHGLEVPGEVDGGPQGIIPPGGKRSVTLNVDQPAATCWFHPHQHGKTGRQVAMGLAGLVVIEDDEILKLMLPKQWGIDDVPVIVQDKKFSADGQIDYQ LDVMTAAVGWFGDTLLTNGAIYPQHAAPRGWLRLRLLNGCNARSLNFATSDNRPLYVIASDGGLLPEPVKVSELPVLMGERFEVLVEVNDNKPFDLVTLPVSQMGMAIA PFDKPHPVMRIQPIAISASGALPDTLSSLPALPSLEGLTVRKLQLSMDPMLDMMGMQMLMEKYGDQAMAGMDHSQMMGHMGHGNMNHMNHGGKFDFHHANKINGQ AFDMNKPMFAAAKGQYERWVISGVGDMMLHPFHIHGTQFRILSENGKPPAAHRAGWKDTVKVEGNVSEVLVKFNHDAPKEHAYMAHCHLLEHEDTGMMLGFTV</p> | ||
| + | |||
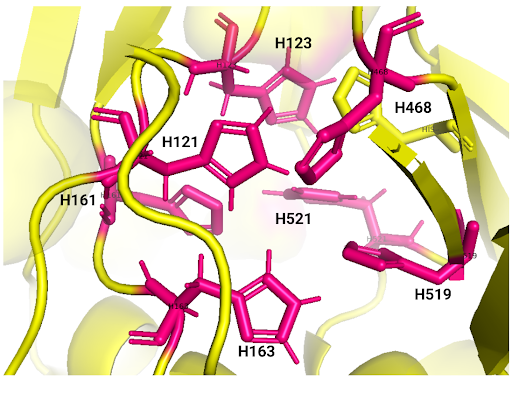
| + | <br><strong>Amino acid active site.</strong></br> | ||
| + | <br><strong>Table I.</strong> Laccase active site amino acids reported by iTASSER.</br> | ||
| + | <br> | ||
| + | |||
| + | <table border="1"> | ||
| + | <tr style="background-color: rgb(236, 167, 233);"><td><strong><center>Amino acid</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H121</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H123</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H161</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H466</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H468</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H519</center></strong></td></tr> | ||
| + | <tr><td><strong><center>H521</center></strong></td></tr> | ||
| + | </table> | ||
| + | |||
| + | <center><img style="vertical-align: bottom;)" width=25% src="https://static.igem.org/mediawiki/parts/0/07/K729002_TecCem_Fig4.png"></center> | ||
| + | <center><strong>Fig. 4</strong> Amino acids of the active site of laccase. </center></strong> | ||
| + | <br></br> | ||
| + | <center><img style="vertical-align: bottom;)" width=25% src="https://static.igem.org/mediawiki/parts/f/f0/K729002_TecCem_Fig5.png"></center> | ||
| + | <center> <strong>Fig. 5</strong> Image focused on the histidines that are part of the active site. </center> | ||
| + | <center> Visualization made in PYMOL C-score obtained from 0.43. </center> | ||
| + | <br><strong>Disulfide bonds</strong></br> | ||
| + | <p align = "justify">Laccase has 3 cysteines, the characteristics are not externalized. These are: CYS158, CYS259, and CYS520.</p> | ||
| + | <br> | ||
| + | <center><img style="vertical-align: bottom;)" width=30% src="https://static.igem.org/mediawiki/parts/2/26/K729002_TecCem_Fig10.png"></center> | ||
| + | </br> | ||
| + | <center><strong>Fig. 10</strong> Laccase cysteines are shown. </center></strong> | ||
| + | <br></br> | ||
| + | <H3>References</H3> | ||
| + | |||
| + | <p align = "justify">[1]<a href="https://benchling.com/brendajmz98/f/lib_ExjL1FRA-laccase/prtn_DvC2wcXk-lacassa_t-ver_proteina/edit"> https://benchling.com/brendajmz98/f/lib_ExjL1FRA-laccase/prtn_DvC2wcXk-lacassa_t-ver_proteina/edit</a></p> | ||
| + | |||
| + | <p align = "justify">[2] Madzak, C., Mimmi, M. C., Caminade, E., Brault, A., Baumberger, S., Briozzo, P., … Jolivalt, C. (2005). <em> Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Engineering, Design and Selection 19(2), 77–84. </em> doi:10.1093/protein/gzj004 </p> | ||
| + | <p align = "justify">[3] Hongyan, L., Zexiong, Z., Shiwei, X., He, X., Yinian, Z., Haiyun, L., & Zhongsheng, Y. (2019). <em> Study on transformation and degradation of bisphenol A by Trametes versicolor laccase and simulation of molecular docking. Chemosphere.</em> doi:10.1016/j.chemosphere.2019.02.14</p> | ||
| + | </html> | ||
| + | |||
| + | =iGEM TecCEM 2022 contribution= | ||
| + | ===Design=== | ||
| + | |||
| + | iGEM TecCEM 2022 Team designed this composite part with coding sequence for Laccase from Trametes versicolor, making a contribution to the part registered by iGEM12_UCL_London, BBa_K729002, even though that laccase was analyzed to degrade polyethylene, but due to being capable of degrading phenolic compounds it is of interest for degrading Endocrine Chemical Disruptors, the problematic set for this year competition by our team, we aim to oxidize EDCs such as BPA into less aggressive, photosensitive removable compounds. The chosen biobrick has been characterized by other teams. Laccase from white rot fungi Trametes versicolor with signal peptide ompA and histidine tag (Protein coding) for endocrine disruptor chemicals in wastewater is a composite part built to produce laccase in the periplasmic space of E.Coli in where proteins are better solubilized[1]. The laccase from Trametes Versicolor was chosen because its wide range of substrates and its ability to carry out its redox reaction in the absence of hydrogen peroxide[2]. Amino acids were optimized for E.Coli as biofabric, because the original sequence comes from Trametes Versicolor. We added both a promoter and a terminator sequence to the construct as it is visible in '''Fig. 1''' in order to allow protein expression using a cloning and expression plasmid for bacterial transformation. | ||
| + | |||
| + | [[File:seq_lactv.png|700px|]] | ||
| + | |||
| + | '''Figure 1'''. Construct sequence design. | ||
| + | |||
| + | When encountered with a susbtrate that have a considerable affinity with its active site, refer to '''Fig. 2''', the laccase oxidizes the phenolic compounds in bioproducts easier to deal with. | ||
| + | [[File:T--TecCEM--ActiveSite.png|thumb|center|250px|<i><b>Figure.2:</b> Active site with histidines. Observed by PYMOL]] | ||
| + | |||
| + | '''pH stability''' | ||
| + | Using Heatmap tool of Protein-sol and the model obtain in PDB of the protein. Based on the heatmap ('''Fig.3'''), its optimal pH is between 5.0 and 8.0, according to the green coloration. | ||
| + | |||
| + | [[File:T--TecCEM--pHStability.png|500px|<i><b>Figure.3:</b> Energy heatmap ( J per aa)</i>]] | ||
| + | |||
| + | '''Hydrophobic and hydrophilic residues''' | ||
| + | Finally, hydrophobic residues of the aminoacids structure of laccase are shown. This prediction was obtained thanks to the ProtScale bioinformatic tool from Expasy [7]. Results show hydrophobic amino acids as positive values and hydrophilic residues as negative values. The most hydrophobic amino acids are found in the 0-20 and 170-180 region and the most hydrophilic in the 160-170 region, '''Fig.4'''. | ||
| + | |||
| + | [[File:Hydrophobicity.gif|thumb|center|250px|<i><b>Figure.4:</b> Hydrophobic and hydrophylic residues</i>]] | ||
| + | |||
| + | |||
| + | ===Characterization: protein modeling and molecular docking=== | ||
| + | |||
| + | [[File:Cbz-laccase.jpeg|thumb|center|250px|<i><b>Figure.5:</b> Superficial interactions between laccase and CBZ</i>]] | ||
| + | |||
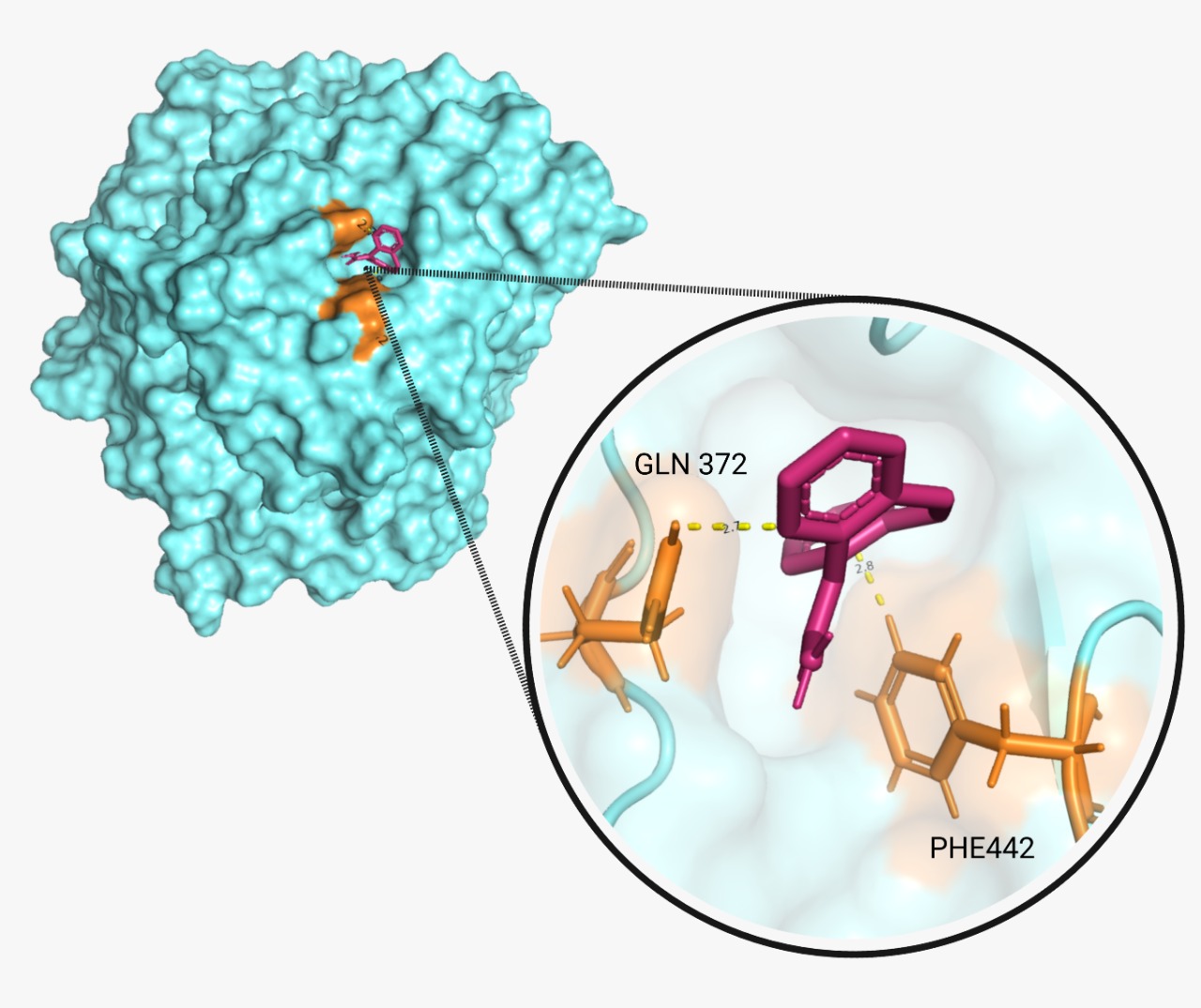
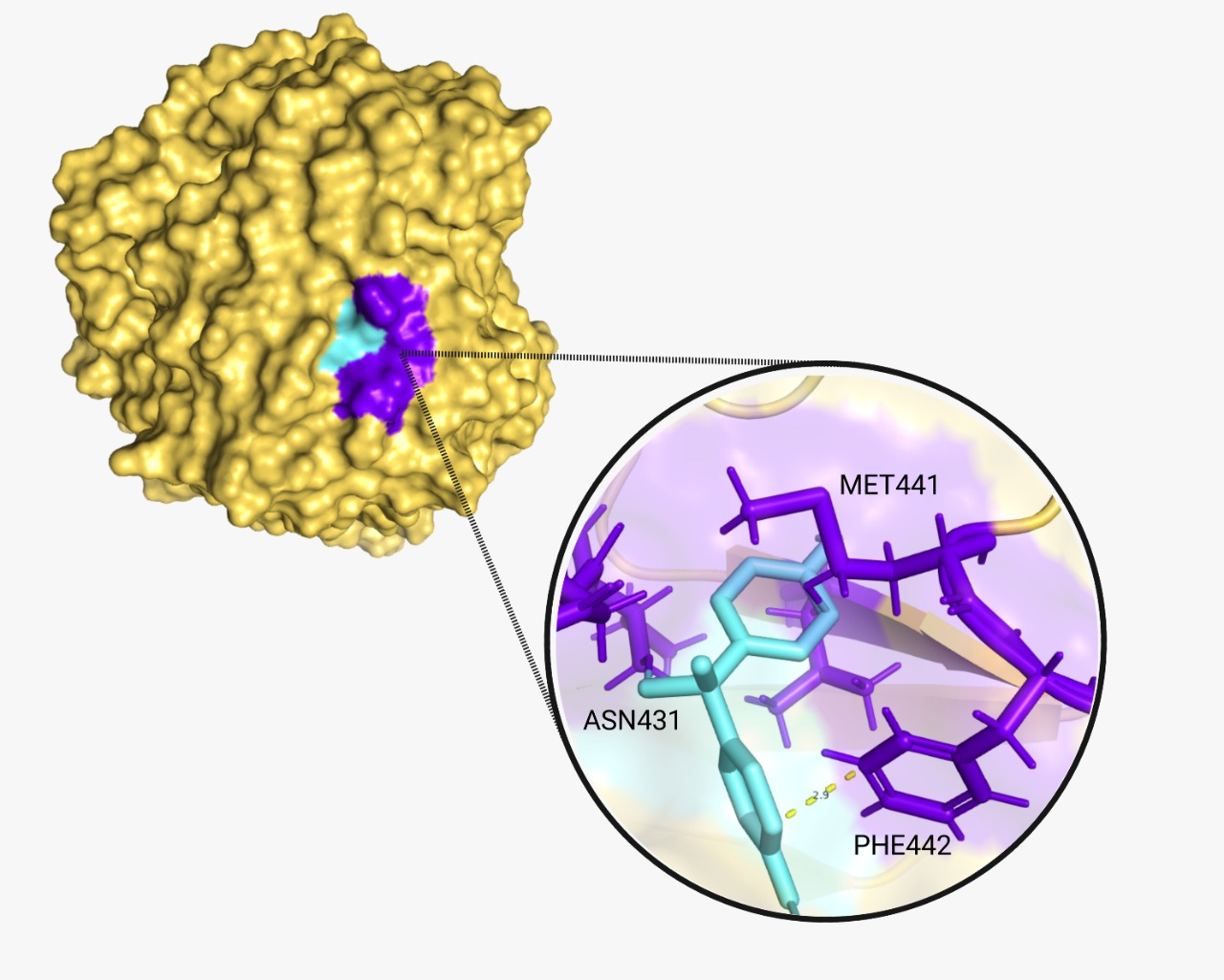
| + | The proper molecular docking was realized with the program AutoDock Vina and Chimera. We obtained a score of -6.1 and -6.3 respectively. These two values demonstrate that the reaction is spontaneous and will occur. A hydrogen bond was encountered between BPA and laccase. Additionally, we identified the amino acids (A.A.) that play a key role in the binding interaction. A tool from Hamburgs University was used for this purpose, called “Pose View”. The program showed that for CBZ ('''Fig.5'''), the key A.A. are glutamine 372 and phenylalanine 442,; and for BPA ('''Fig.6''') are methionine 441, asparagine 431 and phenylalanine 442 again. | ||
| + | |||
| + | [[File:bpa-laccase.jpeg|thumb|center|250px|<i><b>Figure.6:</b> Superficial interactions between laccase and BPA | ||
| + | </i>]] | ||
| + | |||
| + | |||
| + | ===General laccase contribution for His Tag=== | ||
| + | |||
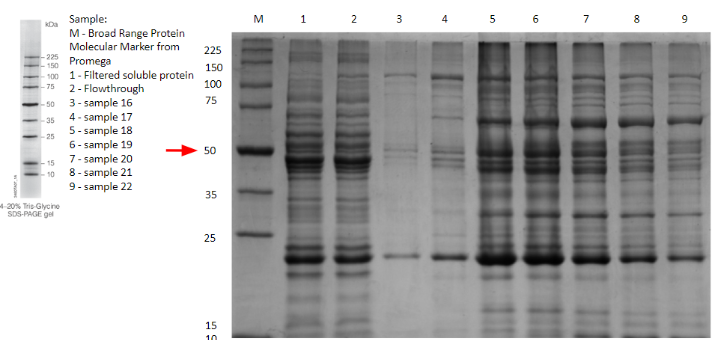
| + | Besides the use of a signal peptide to export the protein into the periplasmic space, the contribution made for general laccases is about the interaction laccase has with nickel. It is reported that laccase interacts, degrades and removes heavy metals and salt metals [3][4]. Nickel is of relevance because the Histidine tag added at the end of recombinant proteins, ideally allows for a nickel chromatography purification of the former. While experimenting, attempts of expression of different laccases found in iGEM registry were done, to determine the ideal for degrading Endocrine Disruptor Chemicals. Various candidates were fusioned with the tag and purified with Ni-NTA, there was one specially from Thermus Thermophilus that between the SDS-Page done for expression assay and the purification with nickel column a observable behavior through the chromatograms was found. | ||
| + | When analyzing IMAC chromatography, it was found that the laccase had bonded to nickel through its active site, compromising the resin’s integrity and forming enzyme-substrate complexes that made the protein appear bigger, as evidenced by the thick bands at around 70 kDa present in lanes 5-9 on gel 1, brand new bands that aren’t visible on the flowthrough nor the solution before purification, which are lanes 2 and 1, respectively ('''Fig.11'''.). | ||
| + | |||
| + | [[File:sds-nic.png|thumb|center|250px|<i><b>Figure.11:</b> SDS-PAGE of laccase 15A’s purified soluble protein utilizing IMAC</i>]] | ||
| + | |||
| + | Despite the intention of implementing a nickel based purification method, with the information found and the observations done during the experiments, it is better for other iGEM teams to opt for different chromatography for protein purification such as ion-exchangable or else. | ||
| + | |||
| + | ===Contribution by SVCE-Chennai 2023=== | ||
| + | |||
| + | The three dimensional structure of the protein was predicted using SWISS-MODEL. The model was validated using Ramachandran plot analysis. It was observed that 95.91 % of the residues were under the favoured region. This confirms the reliability of the predicted protein model <b>Figure 12</b>. | ||
| + | |||
| + | |||
| + | <p> | ||
| + | <html><img src="https://static.igem.wiki/teams/4864/wiki/contribution/bba-k4864008-rc-plot.jpg" alt="Figure 12: Ramachandran Plot for the modeled protein structure" width="500"></html> | ||
| + | </p> | ||
| + | <b>Figure 12: Ramachandran Plot for the modeled protein structure</b> | ||
| + | |||
| + | The molecular docking analysis was performed for laccase and PE (Polyethylene n=10) using AUTODOCK tools. From the 2D (<b>Figure 13</b>) and 3D (<b>Figure 14</b>) interaction maps it was observed that the PE was within the active site pocket and residues LYS474, PRO274, VAL 300, LEU276 was found to interact with the PE substrate. | ||
| + | |||
| + | <p> | ||
| + | <html><img src="https://static.igem.wiki/teams/4864/wiki/contribution/pe-l-2d-interaction.png" alt="Figure 13: 2D interaction map for laccase and polyethylene" width="500"></html> | ||
| + | </p> | ||
| + | <b>Figure 13: 2D interaction map for laccase and polyethylene</b> | ||
| + | |||
| + | <p> | ||
| + | <html><img src="https://static.igem.wiki/teams/4864/wiki/contribution/pe-l-3d-interaction.png" alt="Figure 14: 3D interaction map for laccase and polyethylene" width="500"></html> | ||
| + | </p> | ||
| + | <b>Figure 14: 3D interaction map for laccase and polyethylene</b> | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | <small> | ||
| + | [1] Goulas T, Cuppari A, Garcia-Castellanos R, Snipas S, Glockshuber R, Arolas JL, et al. (2014) The pCri System: A Vector Collection for Recombinant Protein Expression and Purification. PLoS ONE 9(11): e112643.https://doi.org/10.1371/journal.pone.0112643 | ||
| + | [2] Pezzella, C., Macellaro, G., Sannia, G., Raganati, F., Olivieri, G., Marzocchella, A., ... & Piscitelli, A. (2017). Exploitation of Trametes versicolor for bioremediation of endocrine disrupting chemicals in bioreactors. PloS one, 12(6), e0178758. | ||
| + | [3] Ahmadi Khozani, M., Emtiazi, G., Aghaei, S. S., Ghasemi, S. M., & Zolfaghari, M. R. (2021). Application of fungal laccase for heavy metals precipitation using tannin as a natural mediator. International Journal of Environmental Science and Technology, 18(8), 2335-2344. | ||
| + | [4] Enayatizamir, N., Liu, J., Wang, L., Lin, X., & Fu, P. (2020). Coupling Laccase production from Trametes pubescence with heavy metal removal for Economic Waste Water Treatment. Journal of Water Process Engineering, 37, 101357. | ||
Latest revision as of 10:07, 12 October 2023
Laccase for Polyethylene Degradation
Laccase for the degradation of polyethylene and organic pollutants. Sequence pending.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 225
- 1000COMPATIBLE WITH RFC[1000]
Description
The laccase enzyme has been shown to degrade polyethylene when being expressed by a number of different bacteria and fungi. This is due to conserved copper binding sites which couple the oxidation of a substrate with the cleavage of dioxygen bonds, leading to the capability to degrade plastics (particularly polyethylene).
By driving the laccase production using a strong constitutive promoter, we expect overexpression of the protein which will allow it to be secreted into the extracellular medium where it can act upon the target plastic.
Characterisation
Plastic degradation is mediated via a laccase protein. As such, we will be using an enzymatic activity assay to determine that the laccase enzyme is expressed. Laccase catalyses the oxidation of syringaldazine, a reaction that exhibits an observable OD change at 530nm. A sample of syringaldazine can be used as a blank in a spectrophotometer, against a sample containing syringaldazine and our laccase sample, allowing the rate of oxidation to be measured, and hence the enzymatic activity of laccase.
In order to determine the effectiveness of laccase in degrading plastic, we will expose strips of various types of plastic to the laccase expressing bacteria, before viewing the strips under a scanning electron microscope. This will allow us to compare the pitting in plastic samples treated with laccase, to the untreated samples, allowing us to determine the extent of plastic degradation.
Results
Our results indicate a significantly higher rate of oxidation from the cells containing our BioBrick than the control cell line. This indicates that our transformed E. Coli have successfully produced laccase, and in significant enough quantities that it is released into the extracellular space. This allows it to oxidise the syringaldazine utilised in the laccase assay.
The Scanning Electron Microscope (SEM) images below show the effects of subjecting Low Density Polyethylene (LDPE) to three different sets of conditions in order to visualise the degradative properties of cells containing our Laccase construct (BBa_K729006). We can then also compare this with the ability of untransformed E. coli W3110 and mechanical shear in water to breakdown the LDPE target.
All samples were in suspension at 37°C and 200rpm in an incubated shaker for 3 days.
Low magnification (x100) images were used to observe the macro-trend in zonal enzymatic activity. There appears to be increased ‘roughness’ (shown by the small protrusions on the surface as well as areas of darker shading, which represent areas of greater contour), across the surface of the plastic as we progress from the sample treated with water to untransformed E. coli W3110 to E. coli transformed with the UCL Laccase construct.
High magnification (x10 000) images were taken to give more high resolution information regarding the activity of the enzyme on a much smaller scale. The SEM images highlight increased amounts of ‘pitting’ (the generation of small holes in the plastic due to it’s structural breakdown), as we go from water to E. coli W3110 to bacteria containing BBa_K729006. These pits are shown by the much darker areas. This small scale breakdown has a cumulative effect which results in the overall trend seen at the lower magnification.
In conclusion, this collection of SEM images indicates an ability both of the BBa_K729006 transformed cells to produce the Laccase enzyme at greater titres than untransformed bacteria (though the E. coli W3110 is still seen to have some degradative effect), and of the Laccase produced to degrade the polyethylene target.
| Water | W3110 | Laccase | |
|---|---|---|---|
| Low Magnification (100x) |  |
 |

|
| High Magnification (10 000x) | 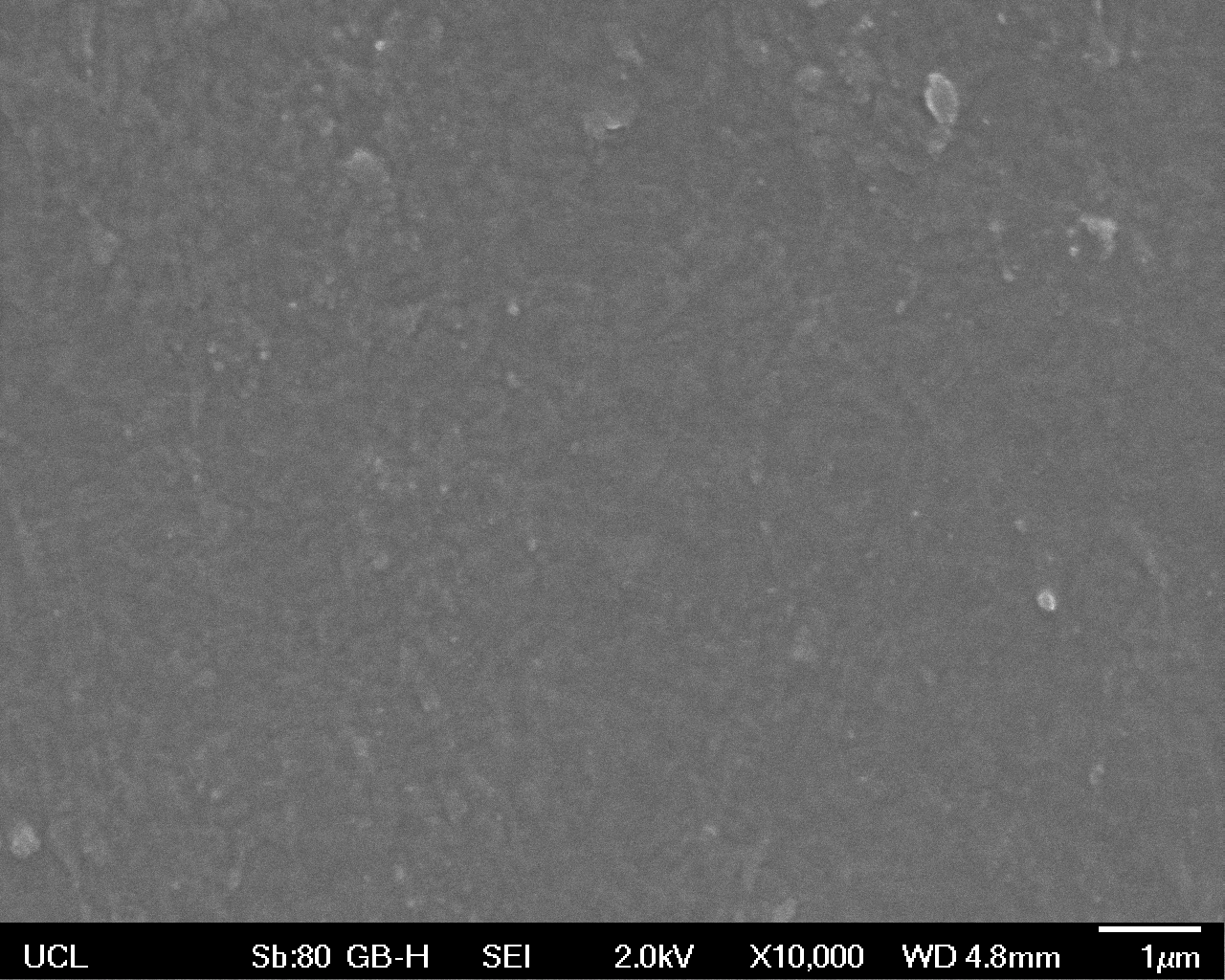 |
 |

|
Modelling
Conclusion
Through the use of a simple quantitative assay, it has been ascertained that E. coli, transformed with our construct has a significantly increased extracellular laccase activity when compared with a control cell line. Combined with the results from the scanning electron microscope, which indicate surface degradation of polyethylene in a relatively short timespan, indicates that this BioBrick holds powerful potential for the break down of polyethylene.
iGEM MITADTBIO_Pune 2019 characterization
Description
Laccase enzyme acts on many different substrates, with its activity also prominent in helping break down polyethylene. Since our aim is to degrade plastic present in menstrual sanitary napkins, it was important to know whether it's activity is affected in the presence of blood. Laccase has mutiple copper ions in its cofactor and therefore it was crucial for us to know whether the activity of laccase is hindered in the presence of other ions present in blood.
Cloning of laccase gene from BBa K729002 into pET28a+
Cloning of laccase in iGEM backbone plasmid will lead to normal low level of laccase expression which will show in vitro activity, but the activity and expressive rate is non-obvious. For the above reasons, we decided to clone laccase gene is pET28a+ expression vector for sustainable production of laccase.
To insert laccase gene into cloning expression vector (pET 28a+), we first designed prefix and suffix primers which contain EcoRI and PstI restriction enzyme sites respectively. The PCR amplification of laccase gene from BBa_K729002 using EcoRI-prefix and PstI-suffix primers give an expected 1.5 kb DNA band as shown in (Fig. 1).
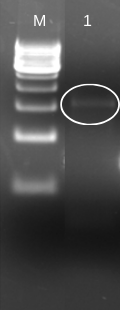
The purified PCR amplified products were restriction double digested with EcoRI-PstI, and ligated at same sites in pET28a+ (5.3 kb) expression vector and transformed recombinant plasmid into E. coli (JM109). The transformation result was shown in Figure 2. To check whether the construction was successful, we picked few colonies to extract their plasmids. The inclusion of insert into the recombinant plasmid was verified both by restriction digestion.
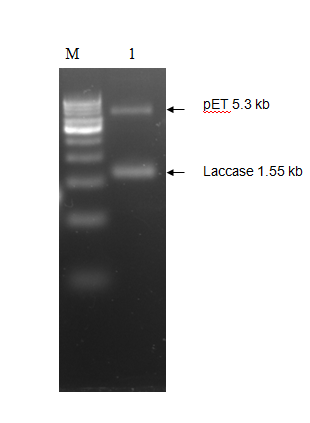
We tested the expression of laccase enzyme by transforming the pET28a+ recombinant carrying pET-laccase gene into E. coli expression strain BL21. To overexpress laccase enzyme, E. coli expression strain BL21 containing laccase gene under IPTG-inducible T7 promoter was induced by 1mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3hrs and 18 hrs at 37°C, corresponding proteins were separated by 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue G250 (CBB). Prior to induction with IPTG, expression of laccase is not detectable by SDS-PAGE (Figure 3 Lane 1). After IPTG induction recombinant, protein bands of about 58 kDa was observed which is in agreement with the theoretical molecular weight of laccase which is 58.87 kDa (Figure 3 Lane 2).
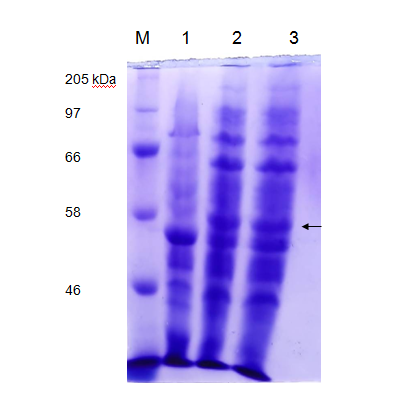
In vitro laccase assay
We tested whether expressed laccase is active through enzyme activity experiments. To detect laccase enzyme activity, colorimetric assay was carried out using 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) or ABTS method. The nonphenolic dye ABTS is oxidized by laccase into water soluble chromogen ABTS+ which is more stable and preferred state of the cation radical. ABTS+ can be monitored spectrophotometrically at 420 nm. The concentration of the cation radical responsible for the intense blue-green color can be correlated to enzyme activity and is read at 420nm. The supernatant of overnight grown bacteria culture acted as a source of laccase enzyme. The enzyme activity experiment was carried out by using 100 μL of enzyme and 100 μL of ABTS solution (0. 5 mM) in sodium acetate buffer (pH 5). The enzymatic units (U) defined as the amount of enzyme transforming 1 µmol of substrate per minute is calculated by the following formula: Equation : U/L = (∆E×Vt)/(ε×d×Vs)
with ∆E being the change in the extinction of light [min-1] at 420 nm
ε being the molar absorption coefficient of ABTS [M-1 cm-1]
d being the layer thickness [cm] in your cell that the light has to pass
Vt is the total volume measured and Vs is the volume of the enzyme stock solution added to the ABTS stock solution.
It is demonstrated in the given graph by in vitro ABTS assay that recombinant laccase protein can oxidize ABTS. It is essential for our project that recombinant microorganisms should be capable of polyethylene deterioration in the presence of blood and/or dried blood. To check the effect of blood on laccase enzyme activity, we performed the ABTS assay in the presence of different concentrations of blood.
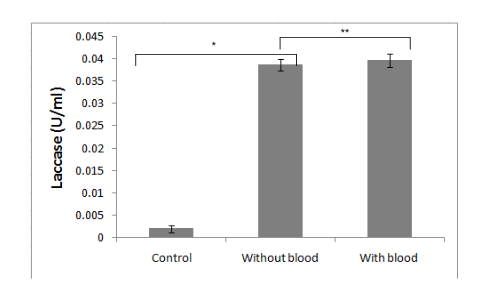
Conclusion
It was clearly observed that the laccase activity is not significantly affected by blood.
Added by LZU-HS-China-C
We found that the laccase BBa_K729002 may also have the effect of degrading antibiotics. So we used this laccase to complete the verification experiment of degrading antibiotics.
At the same time, in order to enhance its function, we added INP protein to the N segment of BBa_K729002, so that the laccase was anchored on the surface of the bacteria through cell surface display technology.
In order to facilitate the comparison of the degradation ability of BBa_K729002 and BBa_K4332002 with INP added, we will compare and show these two parts.
(Correspondence between the gene in the picture and the gene in part:BBa_K729002----lacc6、BBa_K4332002----IL)
Usage and Biology
Laccase is an oxidoreductase that uses oxygen as an electron acceptor. It can oxidize a variety of aromatic compounds and produce water as a by-product. In the 19th century, humans first discovered laccase in Japanese sumac. Because of its outstanding application value in biotechnology applications, scientists have gradually deepened their research on laccase. They discovered that laccase is also present in animals and microorganisms. Laccases derived from different hosts have different functions in the body. Among them, fungal laccases derived from white rot fungi, Trametes, Pleurotus ostreatus and other fungal laccases occupy the mainstream of the current laccase application process. This type of laccase has a relatively wide range of substrate specificity during the application process, and does not require hydrogen peroxide. With the participation of, a variety of phenolic compounds and their derivatives can be directly oxidized with oxygen as the electron acceptor.
In recent years, the main methods for removing antibiotic residues from different sources are traditional methods such as high-temperature composting, fermentation; oxidation, adsorption, electrochemical treatment, membrane method, and microbial degradation. However, these methods have the disadvantages of low removal efficiency, high cost of use, and the possibility of secondary pollution to the environment. Moreover, these methods cannot fundamentally solve the problem of antibiotic residues in poultry. Therefore, it is necessary to find a direct, effective and low-cost method to remove antibiotic residues in poultry. At present, many studies have shown that fungal laccase plays an important role in the process of treating antibiotic residues in wastewater. Three laccase-producing genes LACC6, LACC9, and LACC10 obtained from Pleurotus ostreatus were heterologously expressed in Pichia pastoris and found that the efficiency of degrading sulfa antibiotics reached more than 97%.
Heterologous expression and application advantages of fungal laccase
Studies have shown that fungal laccase is a multi-copper oxidase that can deliver 4 copper ions at the same time, so it can catalyze a variety of phenolic compounds and aromatic compounds. Based on its wide range of substrates, laccase has become a useful biocatalyst in the application of biotechnology, especially in the field of bioremediation, including the degradation of lignin, the decolorization of synthetic dye wastewater, and the degradation and detoxification of environmental pollutants. However, the laccase secreted by wild strains is limited in yield and difficult to purify, on the other hand, its activity is susceptible to environmental factors. In recent years, microbial fermentation engineering has gradually emerged, and the fermentation production of most laccases mainly stays at the level of shake flasks and fermentation tanks. Ryan et al. used an air-lift fermentor to ferment Trametes villus, and the laccase activity in the fermentation broth could reach 11.8 U/ml. At this stage, although the production and application of some fungal laccases can be realized by using the fermentation process, the culture conditions of microorganisms such as temperature, pH, humidity, and the length of the fermentation cycle will affect the production of laccase during the fermentation process. Therefore, most laccases are fermented The process only stays at the laboratory scale. Based on the above shortcomings, it is very important to develop an efficient, low-cost, and environmentally friendly method for producing laccase.
At this stage, researchers have used molecular biology methods to heterologously express laccase genes on the basis of overcoming many shortcomings of traditional laccase purification processes and fermentation processes, in order to obtain laccase with higher yield and enzymatic activity. At present, according to production requirements, laccases from different sources have been successfully expressed in various host cells such as bacteria, fungi, and insect rod cells. At present, in industrial applications, fungal cells such as Pichia pastoris, Saccharomyces cerevisiae and Kluyveromyces lactis dominate the heterologous expression process of laccase. These fungal cells have the following advantages in the application process: first, high-density fermentation can increase the production of laccase, second, the engineered strain has higher enzyme activity, and third, most fungal host cells have been confirmed to be directly edible by humans.
In recent years, with the deepening of research, scientists have found that laccase also has many shortcomings in fungal heterologous expression. When the engineered strain is fermented, it is found that the fermentation cycle is longer and usually takes about one week, and the strain culture consumes a lot. In their research, Maestre et al. found that host yeast can inactivate laccase by interfering with the glycosylation metabolism of laccase. Laccase-producing bacteria from the same source often have multiple laccase genes at the same time, and the existence of these genes ultimately leads to a more complex and diverse isozyme system of laccase. Therefore, when the researchers performed heterologous expression of these isoenzymes, they found that not all fungal expression systems can maximize the productivity of laccase, and the laccase activity produced by heterologous expression in yeast cells is far less than that of wild-type laccase. Strains are also susceptible to factors such as pH. Compared with wild strains, the half-life of laccase in some expression systems has also changed. Comprehensive analysis of the heterologous expression of laccase in fungi, the bacterial expression system has been gradually applied to the direction of heterologous expression of laccase due to its easy operation, short culture period, and strong reproductive ability.
The ability of fungal laccase to degrade sulfadiazine
EcN-Lacc6 refers to the fungal laccase gene expressed in the cell
This experiment mainly uses HPLC to quantitatively analyze the residues of sulfadiazine. The process involves two parts: sample pretreatment and HPLC on-machine detection. The specific experimental method is as follows;
(1) Sample pretreatment. First, EcN-IL and the control wild-type EcN, the unloaded EcN and the intracellular expression strain EcN-Lacc6 were inoculated into LB medium at 37°C overnight at an inoculum of 1%. After centrifugation to remove the supernatant, the bacteria were collected (6000 rpm, 10min). The bacteria were washed twice with the same amount of PBS (pH=7.0) buffer solution and prepared into a bacterial suspension with a cell concentration of 1.0 × 109 CFU/mL. Take 10 mL of the cell suspension and add SDZ sodium salt with a concentration of 30, 50, and 100 mg/L into it, with three replicates in each group. Incubate at room temperature for 3 hours and then centrifuge (10000 r/min, 10 min), collect the supernatant into a 10mL PE tube, and finally filter it into a special brown sample bottle for HPLC using a 0.22μm filter membrane.
(2) HPLC on-machine detection conditions selection. HPLC loading conditions selection: select VWD detector, detection wavelength is set to 270 nm, methanol and ultrapure water are organic phase, mobile phase: methanol: water=25:75, detection column Waters XTERRA MS C18 column (250× 4.6 mm), the column temperature is set to 30°C, and the mobile phase flow rate is 1.0 mL/min. HPLC detection data is calculated using external standard method.
a, b, c show the degradation rate analysis of EcN-IL to sulfadiazine at different concentrations (30, 50 and 100 mg/L). Compared with the control EcN group, the degradation rate of EcN-IL to SDZ can reach up to 30 ±2.3%.
Contribution: Hong Kong JSS 2021
Usage and Biology
In iGEM competition, 2012 Team UCL London, 2012 Team Bielefeld-Germany, 2018 Team HKUST, and 2019 Team MITADTBIO have explored the use of laccase to degrade Polyethylene plastic (BBa_K729002) and showing the enzymatic property of laccase (BBa_K863005). The teams have demonstrated that the laccase protein can be heterogeneously expressed in E. coli and being secreted to the extracellular spaces.
In our project, we focused on the aflatoxin B1 degrading ability of laccase. From our knowledge, there is no iGEM project addressing the AFB1 degradation ability of laccase yet.
Aflatoxin (AF) is a family of carcinogenic toxins produced by Aspergillus sp.. According to the World Health Organization (WHO), 25% of food crops are destroyed due to aflatoxin contamination each year. About 5 billion people are at risk of chronic AF exposure and more than 80% of them will develop AF-related diseases such as hepatocellular carcinoma and liver failure. [2] Among all AF, aflatoxin B1 (AFB1) is considered the most potent and chronic. [3]
Different groups have demonstrated the effectiveness of AFB1 degradation by using native and recombinant laccase [4]. While Yang, P. et al. 's group in 2021 has demonstrated that recombinant laccase produced from P. pastoris could decrease ~48.5% of AFB1 in peanut samples after 18 hour. (Figure 1)[7] This result further support our plan that a detoxifying spray of laccase can help to tackle the problem of AFB1 contaminated food.
Although there are other enzymes that were found to have AFB1 degrading ability, but they either shown lower efficacy or lack of sufficient studies to prove their function when they are heterogeneously expressed. (Table 2). Thus, laccase is one of the most well-studied target that aroused our interest to study in our project.
Among all the laccase produced by different species reviewed, laccase produced by Trametes versicolor (white rot) was found to be the most effective in AFB1 degradation. [3,5] Thus, our team proposes to heterogeneously express this tvLac in probiotic E. coli and to see if it can be a plausible measure to degrade AFB1 in food and act as a food preservative for Aspergillus sp. infection.
We have decided a basic part coding for tvLac for our 2021 phase 1 iGEM project. (BBa_K3746002) and expression composite parts of FDR-A (BBa_K3746006), (BBa_K3746007), and (BBa_K3746009).
Reference
[1] Sumathi T, Viswanath B, Sri Lakshmi A, SaiGopal DV. Production of Laccase by Cochliobolus sp. Isolated from Plastic Dumped Soils and Their Ability to Degrade Low Molecular Weight PVC. Biochem Res Int. 2016;2016:9519527. doi: 10.1155/2016/9519527. Epub 2016 May 12. PMID: 27293894; PMCID: PMC4880699.
[2] Organization WH. aflatoxin. Manuf Comput Solut. 2000;6(8):20-3
[3] Okwara, P. C., Afolabi, I. S., & Ahuekwe, E. F. (2021). Application of laccase in aflatoxin B1 degradation: A Review. IOP Conference Series: Materials Science and Engineering, 1107(1), 012178. https://doi.org/10.1088/1757-899x/1107/1/012178
[4] Alberts, J. F., Gelderblom, W. C. A., Botha, A., & van Zyl, W. H. (2009). Degradation of aflatoxin B1 by fungal laccase enzymes. International Journal of Food Microbiology, 135(1), 47–52. https://doi.org/10.1016/j.ijfoodmicro.2009.07.022
[5] Verheecke, C., Liboz, T., & Mathieu, F. (2016). Microbial degradation of aflatoxin B1: Current status and future advances. International Journal of Food Microbiology, 237, 1–9. https://doi.org/10.1016/j.ijfoodmicro.2016.07.028
[6] Guo, Y., Qin, X., Tang, Y., Ma, Q., Zhang, J., & Zhao, L. (2020). Cota Laccase, a novel aflatoxin oxidase from bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chemistry, 325, 126877. https://doi.org/10.1016/j.foodchem.2020.126877
[7] Yang, P., Xiao, W., Lu, S., Jiang, S., Zheng, Z., Zhang, D., Zhang, M., Jiang, S., & Jiang, S. (2021). Recombinant expression of trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) in engineering pichia Pastoris GS115 and application in AFB1 degradation in AFB1-contaminated peanuts. Toxins, 13(5), 349. https://doi.org/10.3390/toxins13050349
Team TecCEM 2022 detailed enzyme description
Laccase of Trametes Versicolor
iGEM Tec CEM 2022’s construct
Summary
The amino acid sequence was entered in the iTASSER platform to know the structures. Thanks to this model and the subsequent visualization in PYMOL, it was possible to identify the amino acids that are part of the active site, the secondary structures that give it form, and the possible formation of disulfide bonds. Likewise, this model was used to evaluate the pHs at which laccase is most stable, and finally the isoelectric point, to determine its charge at different pHs.

Number of amino acids
535
Molecular Weight
58521.33 Da
Isoelectric Point
(pI)6.44
Extinction Coefficient
| Cys fully reduced | 62910.00 M-1cm-1 | |
| Abs 0.1% (1 g/l) | 1.075 | |
| Cys fully oxidized | 63035.00 M-1cm-1 | |
| Abs 0.1% (1 g/l) | 1.077 |
Instability Index
31.54 (stable)
Optimal pH
The PDB file of the modeled laccase was obtained in iTASSER and entered into the Heatmap tool of Protein-sol. Based on the heatmap, its optimal pH is between 5.0 and 8.0, according to the green coloration.

Isoelectric Point
Based on the heatmap, its PI is close to pH 6.9, due to the white coloration and almost zero charges. Both heatmaps were obtained from https://protein-sol.manchester.ac.uk/heatmap

Amino Acids frequencies
| Ala | A | 50 | 9.3% | |||
| Arg | R | 20 | 3.7% | |||
| Asn | N | 21 | 3.9% | |||
| Asp | D | 31 | 5.8% | |||
| Cys | C | 3 | 0.6% | |||
| Gln | Q | 24 | 4.5% | |||
| Glu | E | 21 | 3.9% | |||
| Gly | G | 52 | 9.7% | |||
| His | H | 23 | 4.3% | |||
| Ile | I | 22 | 4.1% | |||
| Leu | L | 56 | 10.4% | |||
| Lys | K | 26 | 4.9% | |||
| Met | M | 31 | 5.8% | |||
| Phe | F | 18 | 3.4% | |||
| Pro | P | 35 | 6.5% | |||
| Ser | S | 21 | 3.9% | |||
| Thr | T | 27 | 5.0% | |||
| Trp | W | 9 | 1.7% | |||
| Tyr | Y | 9 | 1.7% | |||
| Val | V | 37 | 6.9% | |||
| Pyl | O | 0 | 0.0% | |||
| Pyl | U | 0 | 0.0% |
Amino acid sequence:
MKKITAAAGLLLLAAQPAMAMQRRDFLKYSVALGVASALPLWSRAVFAAERPTLPIPDLLTTDARNRIQLTIGAGQSTFGGKTATTWGYNGNLLGPAVKLQRGKAVTVDIY NQLTEETTLHWHGLEVPGEVDGGPQGIIPPGGKRSVTLNVDQPAATCWFHPHQHGKTGRQVAMGLAGLVVIEDDEILKLMLPKQWGIDDVPVIVQDKKFSADGQIDYQ LDVMTAAVGWFGDTLLTNGAIYPQHAAPRGWLRLRLLNGCNARSLNFATSDNRPLYVIASDGGLLPEPVKVSELPVLMGERFEVLVEVNDNKPFDLVTLPVSQMGMAIA PFDKPHPVMRIQPIAISASGALPDTLSSLPALPSLEGLTVRKLQLSMDPMLDMMGMQMLMEKYGDQAMAGMDHSQMMGHMGHGNMNHMNHGGKFDFHHANKINGQ AFDMNKPMFAAAKGQYERWVISGVGDMMLHPFHIHGTQFRILSENGKPPAAHRAGWKDTVKVEGNVSEVLVKFNHDAPKEHAYMAHCHLLEHEDTGMMLGFTV
Amino acid active site.
Table I. Laccase active site amino acids reported by iTASSER.


Disulfide bonds
Laccase has 3 cysteines, the characteristics are not externalized. These are: CYS158, CYS259, and CYS520.

References
[1] https://benchling.com/brendajmz98/f/lib_ExjL1FRA-laccase/prtn_DvC2wcXk-lacassa_t-ver_proteina/edit
[2] Madzak, C., Mimmi, M. C., Caminade, E., Brault, A., Baumberger, S., Briozzo, P., … Jolivalt, C. (2005). Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Engineering, Design and Selection 19(2), 77–84. doi:10.1093/protein/gzj004
[3] Hongyan, L., Zexiong, Z., Shiwei, X., He, X., Yinian, Z., Haiyun, L., & Zhongsheng, Y. (2019). Study on transformation and degradation of bisphenol A by Trametes versicolor laccase and simulation of molecular docking. Chemosphere. doi:10.1016/j.chemosphere.2019.02.14
iGEM TecCEM 2022 contribution
Design
iGEM TecCEM 2022 Team designed this composite part with coding sequence for Laccase from Trametes versicolor, making a contribution to the part registered by iGEM12_UCL_London, BBa_K729002, even though that laccase was analyzed to degrade polyethylene, but due to being capable of degrading phenolic compounds it is of interest for degrading Endocrine Chemical Disruptors, the problematic set for this year competition by our team, we aim to oxidize EDCs such as BPA into less aggressive, photosensitive removable compounds. The chosen biobrick has been characterized by other teams. Laccase from white rot fungi Trametes versicolor with signal peptide ompA and histidine tag (Protein coding) for endocrine disruptor chemicals in wastewater is a composite part built to produce laccase in the periplasmic space of E.Coli in where proteins are better solubilized[1]. The laccase from Trametes Versicolor was chosen because its wide range of substrates and its ability to carry out its redox reaction in the absence of hydrogen peroxide[2]. Amino acids were optimized for E.Coli as biofabric, because the original sequence comes from Trametes Versicolor. We added both a promoter and a terminator sequence to the construct as it is visible in Fig. 1 in order to allow protein expression using a cloning and expression plasmid for bacterial transformation.
Figure 1. Construct sequence design.
When encountered with a susbtrate that have a considerable affinity with its active site, refer to Fig. 2, the laccase oxidizes the phenolic compounds in bioproducts easier to deal with.
pH stability Using Heatmap tool of Protein-sol and the model obtain in PDB of the protein. Based on the heatmap (Fig.3), its optimal pH is between 5.0 and 8.0, according to the green coloration.
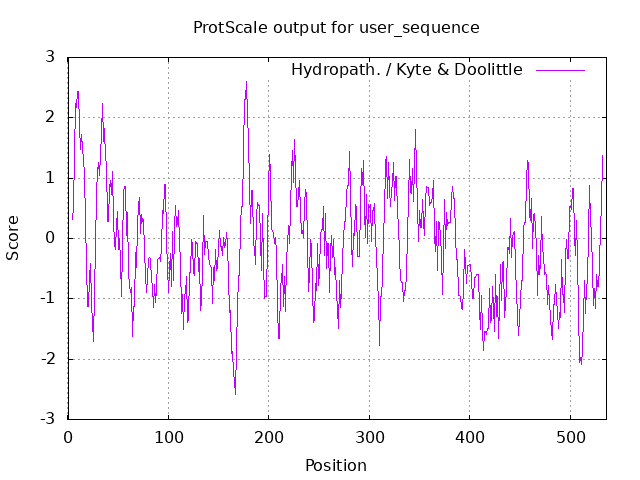
Hydrophobic and hydrophilic residues Finally, hydrophobic residues of the aminoacids structure of laccase are shown. This prediction was obtained thanks to the ProtScale bioinformatic tool from Expasy [7]. Results show hydrophobic amino acids as positive values and hydrophilic residues as negative values. The most hydrophobic amino acids are found in the 0-20 and 170-180 region and the most hydrophilic in the 160-170 region, Fig.4.
Characterization: protein modeling and molecular docking
The proper molecular docking was realized with the program AutoDock Vina and Chimera. We obtained a score of -6.1 and -6.3 respectively. These two values demonstrate that the reaction is spontaneous and will occur. A hydrogen bond was encountered between BPA and laccase. Additionally, we identified the amino acids (A.A.) that play a key role in the binding interaction. A tool from Hamburgs University was used for this purpose, called “Pose View”. The program showed that for CBZ (Fig.5), the key A.A. are glutamine 372 and phenylalanine 442,; and for BPA (Fig.6) are methionine 441, asparagine 431 and phenylalanine 442 again.
General laccase contribution for His Tag
Besides the use of a signal peptide to export the protein into the periplasmic space, the contribution made for general laccases is about the interaction laccase has with nickel. It is reported that laccase interacts, degrades and removes heavy metals and salt metals [3][4]. Nickel is of relevance because the Histidine tag added at the end of recombinant proteins, ideally allows for a nickel chromatography purification of the former. While experimenting, attempts of expression of different laccases found in iGEM registry were done, to determine the ideal for degrading Endocrine Disruptor Chemicals. Various candidates were fusioned with the tag and purified with Ni-NTA, there was one specially from Thermus Thermophilus that between the SDS-Page done for expression assay and the purification with nickel column a observable behavior through the chromatograms was found. When analyzing IMAC chromatography, it was found that the laccase had bonded to nickel through its active site, compromising the resin’s integrity and forming enzyme-substrate complexes that made the protein appear bigger, as evidenced by the thick bands at around 70 kDa present in lanes 5-9 on gel 1, brand new bands that aren’t visible on the flowthrough nor the solution before purification, which are lanes 2 and 1, respectively (Fig.11.).
Despite the intention of implementing a nickel based purification method, with the information found and the observations done during the experiments, it is better for other iGEM teams to opt for different chromatography for protein purification such as ion-exchangable or else.
Contribution by SVCE-Chennai 2023
The three dimensional structure of the protein was predicted using SWISS-MODEL. The model was validated using Ramachandran plot analysis. It was observed that 95.91 % of the residues were under the favoured region. This confirms the reliability of the predicted protein model Figure 12.

Figure 12: Ramachandran Plot for the modeled protein structure
The molecular docking analysis was performed for laccase and PE (Polyethylene n=10) using AUTODOCK tools. From the 2D (Figure 13) and 3D (Figure 14) interaction maps it was observed that the PE was within the active site pocket and residues LYS474, PRO274, VAL 300, LEU276 was found to interact with the PE substrate.

Figure 13: 2D interaction map for laccase and polyethylene

Figure 14: 3D interaction map for laccase and polyethylene
References
[1] Goulas T, Cuppari A, Garcia-Castellanos R, Snipas S, Glockshuber R, Arolas JL, et al. (2014) The pCri System: A Vector Collection for Recombinant Protein Expression and Purification. PLoS ONE 9(11): e112643.https://doi.org/10.1371/journal.pone.0112643 [2] Pezzella, C., Macellaro, G., Sannia, G., Raganati, F., Olivieri, G., Marzocchella, A., ... & Piscitelli, A. (2017). Exploitation of Trametes versicolor for bioremediation of endocrine disrupting chemicals in bioreactors. PloS one, 12(6), e0178758. [3] Ahmadi Khozani, M., Emtiazi, G., Aghaei, S. S., Ghasemi, S. M., & Zolfaghari, M. R. (2021). Application of fungal laccase for heavy metals precipitation using tannin as a natural mediator. International Journal of Environmental Science and Technology, 18(8), 2335-2344. [4] Enayatizamir, N., Liu, J., Wang, L., Lin, X., & Fu, P. (2020). Coupling Laccase production from Trametes pubescence with heavy metal removal for Economic Waste Water Treatment. Journal of Water Process Engineering, 37, 101357.