Part:BBa_K3746002
tvLac-6His
Description
This is a part design for the Phase I project (2021 iGEM) of Team Hong Kong JSS.
This basic part is the coding sequence of laccase gene from fungal species - Trametes versicolor, which was reported to have a high activity in degradation of aflatoxin B1 (AFB1).
Laccase is a multicopper oxidase found in various organisms, such as plants and fungi. Laccase catalyzes the oxidation of phenol groups in aromatic organic substances.
Laccase has a wide range of applications, such as melanin production, lignin degradation, and lacquer synthesis. In recent years, laccase produced by some fungal species has also been reported to be involved in the bio-degradation of polyvinyl chloride (PVC) plastic in laboratory conditions [1]. Thus, it may also be a solution to the problem of micro-plastic waste pollution.
Usage and Biology
In iGEM competition, 2012 Team UCL London, 2012 Team Bielefeld-Germany, 2018 Team HKUST, and 2019 Team MITADTBIO have explored the use of laccase to degrade Polyethylene plastic (BBa_K729002) and showing the enzymatic property of laccase (BBa_K863005). The teams have demonstrated that the laccase protein can be heterogeneously expressed in E. coli and being secreted to the extracellular spaces.
In our project, we focused on the aflatoxin B1 degrading ability of laccase. From our knowledge, there is no iGEM project addressing the AFB1 degradation ability of laccase yet.
Aflatoxin (AF) is a family of carcinogenic toxins produced by Aspergillus sp.. According to the World Health Organization (WHO), 25% of food crops are destroyed due to aflatoxin contamination each year. About 5 billion people are at risk of chronic AF exposure and more than 80% of them will develop AF-related diseases such as hepatocellular carcinoma and liver failure. [2] Among all AF, aflatoxin B1 (AFB1) is considered the most potent and chronic. [3]
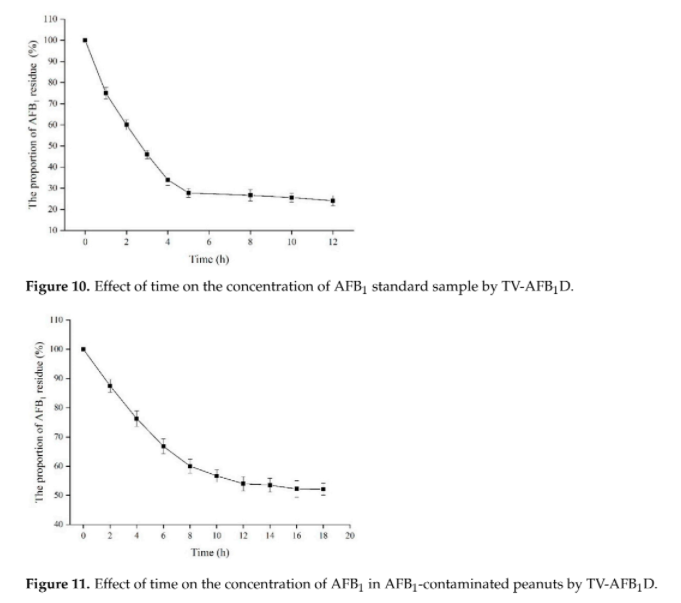
Different groups have demonstrated the effectiveness of AFB1 degradation by using native and recombinant laccase [4]. While Yang, P. et al. 's group in 2021 has demonstrated that recombinant laccase produced from P. pastoris could decrease ~48.5% of AFB1 in peanut samples after 18 hour. (Figure 1)[7] This result further support our plan that a detoxifying spray of laccase can help to tackle the problem of AFB1 contaminated food.
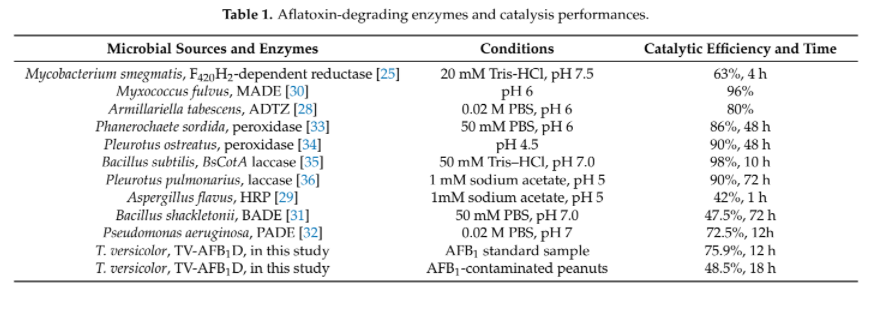
Although there are other enzymes that were found to have AFB1 degrading ability, but they either shown lower efficacy or lack of sufficient studies to prove their function when they are heterogeneously expressed. (Table 2). Thus, laccase is one of the most well-studied target that aroused our interest to study in our project.
Among all the laccase produced by different species reviewed, laccase produced by Trametes versicolor (white rot) was found to be the most effective in AFB1 degradation. [3,5] Thus, our team proposes to heterogeneously express this tvLac in probiotic E. coli and to see if it can be a plausible measure to degrade AFB1 in food and act as a food preservative for Aspergillus sp. infection.
This coding sequence is planned to be used in our composite parts for tvLac enzyme expression (BBa_K3746006), (BBa_K3746007), and (BBa_K3746009).
Note that a C-terminal 6-His Tag (CATCATCATCATCATCAT) was added to this part. For the secretory constructs, (BBa_K3746007) and (BBa_K3746009), the start codon is removed due to the addition of N-terminal signal peptide sequences.
Reference
[1] Sumathi T, Viswanath B, Sri Lakshmi A, SaiGopal DV. Production of Laccase by Cochliobolus sp. Isolated from Plastic Dumped Soils and Their Ability to Degrade Low Molecular Weight PVC. Biochem Res Int. 2016;2016:9519527. doi: 10.1155/2016/9519527. Epub 2016 May 12. PMID: 27293894; PMCID: PMC4880699.
[2] Organization WH. aflatoxin. Manuf Comput Solut. 2000;6(8):20-3
[3] Okwara, P. C., Afolabi, I. S., & Ahuekwe, E. F. (2021). Application of laccase in aflatoxin B1 degradation: A Review. IOP Conference Series: Materials Science and Engineering, 1107(1), 012178. https://doi.org/10.1088/1757-899x/1107/1/012178
[4] Alberts, J. F., Gelderblom, W. C. A., Botha, A., & van Zyl, W. H. (2009). Degradation of aflatoxin B1 by fungal laccase enzymes. International Journal of Food Microbiology, 135(1), 47–52. https://doi.org/10.1016/j.ijfoodmicro.2009.07.022
[5] Verheecke, C., Liboz, T., & Mathieu, F. (2016). Microbial degradation of aflatoxin B1: Current status and future advances. International Journal of Food Microbiology, 237, 1–9. https://doi.org/10.1016/j.ijfoodmicro.2016.07.028
[6] Guo, Y., Qin, X., Tang, Y., Ma, Q., Zhang, J., & Zhao, L. (2020). Cota Laccase, a novel aflatoxin oxidase from bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chemistry, 325, 126877. https://doi.org/10.1016/j.foodchem.2020.126877
[7] Yang, P., Xiao, W., Lu, S., Jiang, S., Zheng, Z., Zhang, D., Zhang, M., Jiang, S., & Jiang, S. (2021). Recombinant expression of trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) in engineering pichia Pastoris GS115 and application in AFB1 degradation in AFB1-contaminated peanuts. Toxins, 13(5), 349. https://doi.org/10.3390/toxins13050349
| None |