Difference between revisions of "Part:BBa K525223"
(→Identification and localisation) |
(→Identification and localisation) |
||
| (20 intermediate revisions by 5 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K525223 short</partinfo> | <partinfo>BBa_K525223 short</partinfo> | ||
| − | [[Image:Bielefeld-Germany2011-S-Layer-Geometrien.jpg|300px|right]] | + | [[Image:Bielefeld-Germany2011-S-Layer-Geometrien.jpg|300px|thumb|right]] |
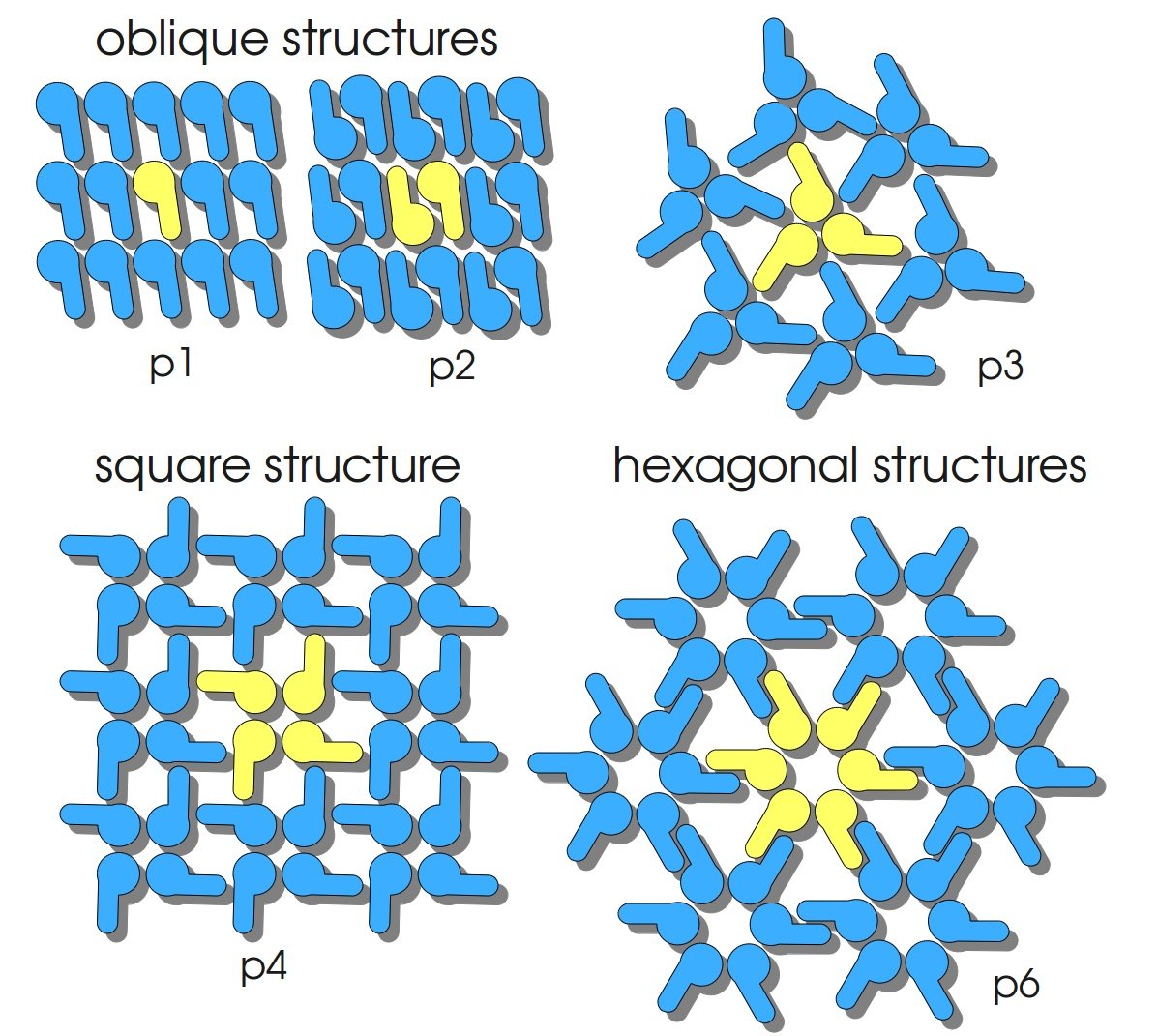
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr ''et al.'', 2007]). | S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr ''et al.'', 2007]). | ||
| Line 19: | Line 19: | ||
!Result | !Result | ||
|- | |- | ||
| − | |rowspan=" | + | |rowspan="4"|[[Part:BBa_K525223#Expression_in_E._coli | Expression (''E. coli'')]] |
| − | + | ||
| − | + | ||
| − | + | ||
|Compatibility | |Compatibility | ||
|''E. coli'' KRX | |''E. coli'' KRX | ||
| Line 34: | Line 31: | ||
|Doubling time (un-/induced) | |Doubling time (un-/induced) | ||
|2.79 h / 7.07 h | |2.79 h / 7.07 h | ||
| + | |- | ||
| + | |rowspan="8"|[[Part:BBa_K525223#Purification_of_SgsE_fusion_protein | Characterization]] | ||
| + | |- | ||
| + | |Number of amino acids | ||
| + | |486 | ||
| + | |- | ||
| + | |Molecular weight | ||
| + | |53.4 kDa | ||
| + | |- | ||
| + | |Theoretical pI | ||
| + | |4.27 | ||
| + | |- | ||
| + | |rowspan="4"|Localization | ||
| + | |cytoplasm | ||
| + | |- | ||
| + | |cell membrane | ||
| + | |- | ||
| + | |partially in culture supernatant | ||
| + | |- | ||
| + | |partially periplasm | ||
|- | |- | ||
|} | |} | ||
| − | </center> | + | </center> |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 50: | Line 67: | ||
<partinfo>BBa_K525223 parameters</partinfo> | <partinfo>BBa_K525223 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
===Expression in ''E. coli''=== | ===Expression in ''E. coli''=== | ||
| − | + | For characterizations, the ''cspB'' gene was fused to a monomeric RFP ([https://parts.igem.org/Part:BBa_E1010 BBa_E1010]) using Gibson assembly. | |
| − | The | + | The CspB|mRFP fusion protein was overexpressed in ''E. coli'' KRX after induction of a T7 polymerase gene in the KRX's genome by supplementation of 0.1 % L-rhamnose using the autinduction protocol developed by Promega. |
| − | [[Image:Bielefeld_2011_CH3_Growthcurve.png|600px|center|thumb| '''Figure 1: | + | [[Image:Bielefeld_2011_CH3_Growthcurve.png|600px|center|thumb| '''Figure 1: Growth curve of ''E. coli'' KRX expressing the fusion protein of CspB and mRFP with and without induction, cultivated at 37 °C in autoinduction medium with and without inductor, respectively. A curve depicting KRX wildtype is shown for comparsion. After autoinduction at approximately 6 h the OD<sub>600</sub> of the induced <partinfo>K525222</partinfo> visibly drops when compared to the uninduced culture. While the induced culture grow significantly slower than KRX wildtype the uninduced seems to be unaffected.''']] |
| − | [[Image:Bielefeld_2011_CH3_RFU_OD.png|600px|center|thumb| '''Figure 2: RFU to OD<sub>600</sub> ratio of ''E. coli'' KRX expressing the fusion protein of CspB and mRFP with and without induction. A curve depicting KRX wildtype is shown for comparsion.''']] | + | [[Image:Bielefeld_2011_CH3_RFU_OD.png|600px|center|thumb| '''Figure 2: RFU to OD<sub>600</sub> ratio of ''E. coli'' KRX expressing the fusion protein of CspB and mRFP with and without induction. A curve depicting KRX wildtype is shown for comparsion. After induction at approximately 6 h the RFU to OD<sub>600</sub> ratio starts to rise in the induced culture. Compared to the uninduced culture the ratio is roughly five times higher. Most likely due to basal transcription the RFU to OD<sub>600</sub> ratio of the uninduced culture starts to rise after 12 hours. The KRX wildtype shows no variation in the RFU to OD<sub>600</sub> ratio.''']] |
===Identification and localisation=== | ===Identification and localisation=== | ||
| − | After a cultivation time of 18 h the mRFP|CspB fusion protein | + | After a cultivation time of 18 h the mRFP|CspB fusion protein was localized in ''E. coli'' KRX. Therefore a part of the produced biomass was mechanically disrupted and the resulting lysate was washed with ddH<sub>2</sub>O. From the other part the periplasm was detached by using an osmotic shock. |
| − | The S-layer fusion protein could not be found in the polyacrylamide gel after a SDS-PAGE of the lysate. This indicated that the fusion protein | + | The S-layer fusion protein could not be found in the polyacrylamide gel after a SDS-PAGE of the lysate. This indicated that the fusion protein integrates into the cell membrane with its lipid anchor. For testing this assumption the washed lysate was treated with ionic, nonionic and zwitterionic detergents to release the mRFP|CspB out of the membranes. |
| − | The existance of flourescence in two of the detergent fractions (10 % SDS and 10 % | + | The existance of flourescence in two of the detergent fractions (10 % SDS and 10 % N-lauroyl sarcosine) and the low fluorescence in the wash fraction confirm the hypothesis of an insertion into the cell membrane (fig. 3). An insertion of these S-layer proteins might stabilize the membrane structure and increase the stability of cells against mechanical and chemical treatment. A stabilization of ''E. coli'' expressing S-layer proteins was described by [http://mic.sgmjournals.org/content/156/12/3584.long Lederer ''et al.'', 2010]. |
An other important fact is, that there is actually mRFP fluorescence measurable in such high concentrated detergent solutions. The S-layer seems to stabilize the biologically active conformation of mRFP. | An other important fact is, that there is actually mRFP fluorescence measurable in such high concentrated detergent solutions. The S-layer seems to stabilize the biologically active conformation of mRFP. | ||
| − | In comparison with the mRFP fusion protein of | + | In comparison with the mRFP fusion protein of [https://parts.igem.org/Part:BBa_K525224 K525224], wich has a TAT-sequence, a minor relative fluorescence in all cultivation and detergent fractions were detected (fig. 3). Together with the decreasing RFU after 9 h of cultivation (fig. 2) this results indicate a postive effect of the TAT-sequence on the protein stability. This could be due to a digestion of [https://parts.igem.org/Part:BBa_K525223 K525223] by proteases in the cytoplasm. |
| + | |||
| + | [[Image:Bielefeld 2011 CH3 Purification.png|700px|thumb|center| '''Figure 3: Fluorescence progression of the mRFP[https://parts.igem.org/Part:BBa_E1010 (BBa_E1010)]/CspB fusion protein initiating with the cultivation fractions up to the detergent fractions of the seperate denaturations. Cultivations were carried out in autoinduction medium at 37 ˚C. The cells were mechanically disrupted and the resulting biomass was washed with ddH<sub>2</sub>O and resuspended in the respective detergent. The used detergent acronyms stand for: SDS = 10 % sodium dodecyl sulfate; UTU = 7 M urea and 3 M thiourea; U = 10 M urea; NLS = 10 % n-lauroyl sarcosine; 2 % CHAPS = 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate.''']] | ||
| + | |||
| + | |||
| + | To obtain more specific information about the location of the S-layer fusion protein, after comparison with same treated fraction of ''E. coli'' KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 4. The fusion protein CspB|mRFP [https://parts.igem.org/Part:BBa_E1010 (BBa_E1010)] features a lipid anchor at the carboxy-terminus, but no amino-terminal TAT-sequence. In accordance with other protein variants with and without this features, the protein should be located mainly in the cytoplasm as inclusion bodies or incooperated with its lipid anchor into the cell membrane. Thus, the fraction with 10 % (v/v) SDS as detergent to disintegrate the protein from the cell wall was measured with MALDI-TOF. Results are shown in fig. 4. | ||
| + | |||
| + | |||
| + | [[Image:Bielefeld2011 K525233 Gel2.png|900px|thumb|right| '''Figure 4: MALDI-TOF measurement of CspB/mRFP [https://parts.igem.org/Part:BBa_E1010 (BBa_E1010)] fusion protein in different fractions. Abbreviations are Ma: Marker (PageRuler <sup>TM</sup> Prestained Protein Ladder SM0671), M (medium), PP (periplasm), L (cell lysis with ribolyser), W (wash with ddH<sub>2</sub>O). In the left half of the gel fractions of ''E. coli'' KRX with induced production of fusion protein, the right half shows fractions of ''E. coli'' KRX without carrying the plasmid coding the fusion protein. Colours show the sequence coverage of the gel lane, cutted out of the gel.''']] | ||
| + | |||
| + | Fig. 4 shows, that the protein could be identified in all measured gel bands. The results indicate, that the protein is incorperated into the cell membrane. No fluoresence could be detected in the fractions using urea as detergent (see fig. 3), thus the protein probably does not form inclusion bodies. The fact that there is a comparable low overall fluorescence in the cells is a strong indication that the protein is degraded by ''E. coli'' proteases. The fluorescence in the periplasm and the supernatant is probably due to cell lysis during the periplasm isolation and the cultivation because the measured fluorescence values are comparable low, too. | ||
| + | |||
| + | ===Methods=== | ||
| + | |||
| + | '''Expression of S-layer genes in ''E. coli'' ''' | ||
| + | * Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] | ||
| + | |||
| + | * Medium: LB medium supplemented with 20 mg L<sup>-1</sup> chloramphenicol | ||
| + | ** For autoinduction: Cultivations in LB-medium were supplemented with 0.1 % L-rhamnose as inducer and 0.05 % glucose | ||
| + | |||
| + | |||
| + | '''Measuring of [https://parts.igem.org/Part:BBa_E1010 mRFP]''' | ||
| + | * Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | ||
| + | * Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | ||
| + | * To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination) | ||
| + | * Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings: | ||
| + | ** 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm) | ||
| + | ** Measurement mode: Top | ||
| + | ** Excitation: 584 nm | ||
| + | ** Emission: 620 nm | ||
| + | ** Number of reads: 25 | ||
| + | ** Manual gain: 100 | ||
| + | ** Integration time: 20 µs | ||
| + | |||
| + | |||
| + | '''Tryptic digest of gel lanes for analysis with MALDI-TOF''' | ||
| + | |||
| + | Note: | ||
| + | *Make sure to work under a fume hood. | ||
| + | *Do not work with protective gloves to prevent contamination of your sample with platicizers. | ||
| + | |||
| + | Reaction tubes have to be cleaned with 60% (v/v) CH<sub>3</sub>CN, 0.1% (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing. | ||
| + | |||
| + | *Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices. | ||
| + | *Gel slices should be washed two times. Therefore add 200 µL 30% (v/v) acetonitrile in 0.1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste. | ||
| + | *Dry gel slices at least 30 minutes in a Speedvac. | ||
| + | *Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation. | ||
| + | *Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night. | ||
| + | *Dry gel slices at least 30 minutes in a Speedvac. | ||
| + | *According to the size of the gel slice, add 5 – 20 µL 50% (v/v) ACN / 0,1% (v/v) TFA. | ||
| + | *Samples can be used for MALDI measurement or stored at -20 °C. | ||
| + | |||
| + | Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH<sub>4</sub>HCO<sub>3</sub> | ||
| + | *Therefore solubilize lyophilized Trypsin in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C. | ||
| + | |||
| + | |||
| + | '''Preparation and Spotting for analysis of peptides on Bruker AnchorChips''' | ||
| + | |||
| + | *Spot 0,5 – 1 µL sample aliquot | ||
| + | *Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip. | ||
| + | Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area. | ||
| + | *Dry the prepared spots at room temperature | ||
| + | *Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0,1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial. | ||
| + | *Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots. | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | Lederer FL, Günther TJ, Flemming K, Raff J, Fahmy K, Springer A, Pollmann K (2010) Heterologous expression of the surface-layer-like protein SllB induces the formation of long filaments of Escherichia coli consisting of protein-stabilized outer membrane. [http://mic.sgmjournals.org/content/156/12/3584.long Microbiology. 156(Pt 12):3584-95]. | ||
| + | |||
| + | Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full ''FEMS Microbiol Lett'' 267(2):131-144]. | ||
| − | |||
<!-- --> | <!-- --> | ||
Latest revision as of 11:50, 28 October 2011
S-layer cspB from Corynebacterium halotolerans with lipid anchor, PT7 and RBS
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]).
Usage and Biology
S-layer proteins can be used as scaffold for nanobiotechnological applications and devices by e.g. fusing the S-layer's self-assembly domain to other functional protein domains. It is possible to coat surfaces and liposomes with S-layers. A big advantage of S-layers: after expressing in E. coli and purification, the nanobiotechnological system is cell-free. This enhances the biological security of a device.
Important parameters
| Experiment | Characteristic | Result |
|---|---|---|
| Expression (E. coli) | Compatibility | E. coli KRX |
| Induction of expression | L-rhamnose for induction of T7 polymerase | |
| Specific growth rate (un-/induced) | 0.248 h-1 / 0.098 h-1 | |
| Doubling time (un-/induced) | 2.79 h / 7.07 h | |
| Characterization | ||
| Number of amino acids | 486 | |
| Molecular weight | 53.4 kDa | |
| Theoretical pI | 4.27 | |
| Localization | cytoplasm | |
| cell membrane | ||
| partially in culture supernatant | ||
| partially periplasm |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1102
Illegal XhoI site found at 558 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 225
Illegal NgoMIV site found at 1314
Illegal NgoMIV site found at 1425
Illegal AgeI site found at 216
Illegal AgeI site found at 457
Illegal AgeI site found at 504 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 906
Illegal BsaI.rc site found at 213
Illegal BsaI.rc site found at 591
Illegal BsaI.rc site found at 993
Expression in E. coli
For characterizations, the cspB gene was fused to a monomeric RFP (BBa_E1010) using Gibson assembly.
The CspB|mRFP fusion protein was overexpressed in E. coli KRX after induction of a T7 polymerase gene in the KRX's genome by supplementation of 0.1 % L-rhamnose using the autinduction protocol developed by Promega.
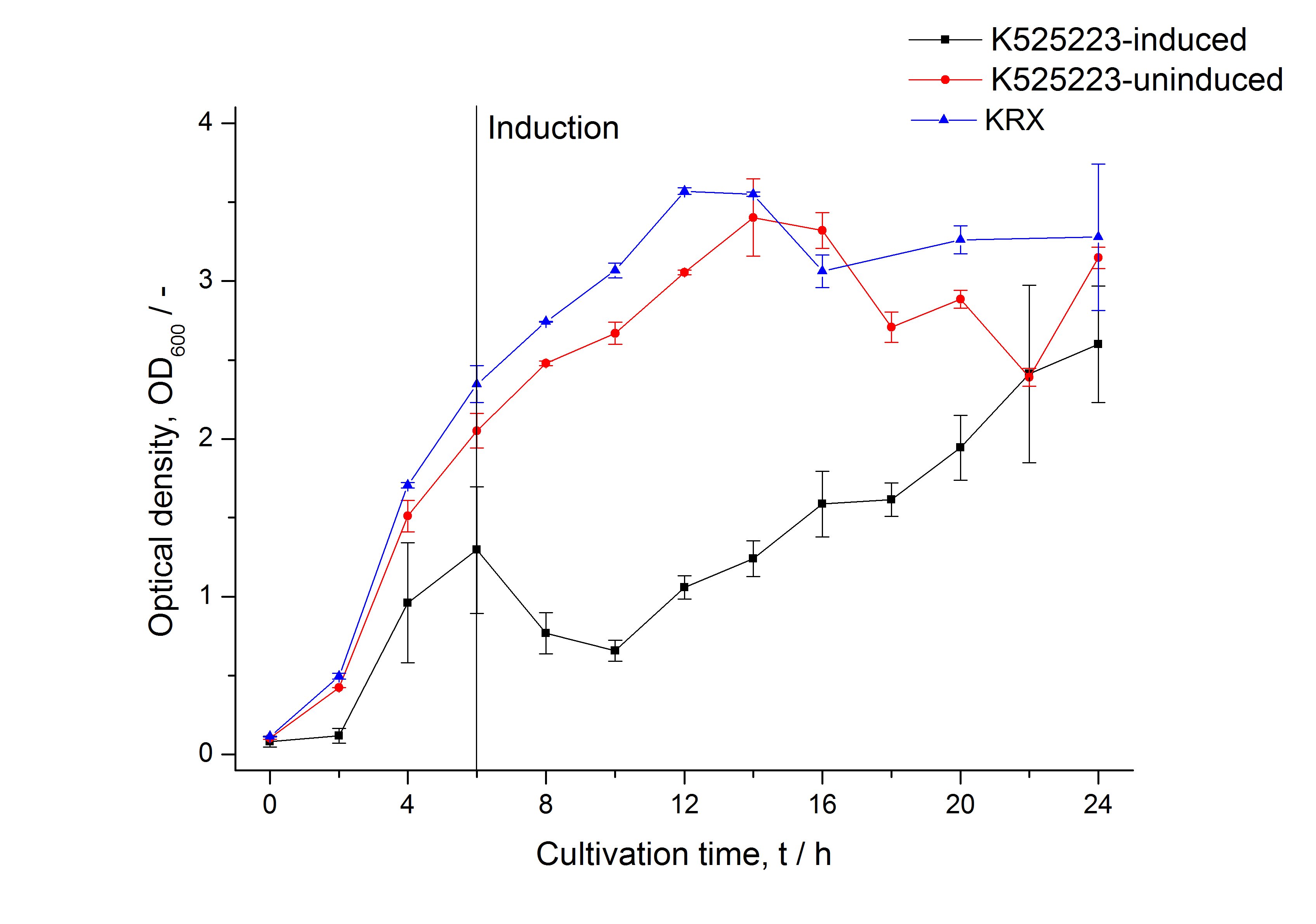
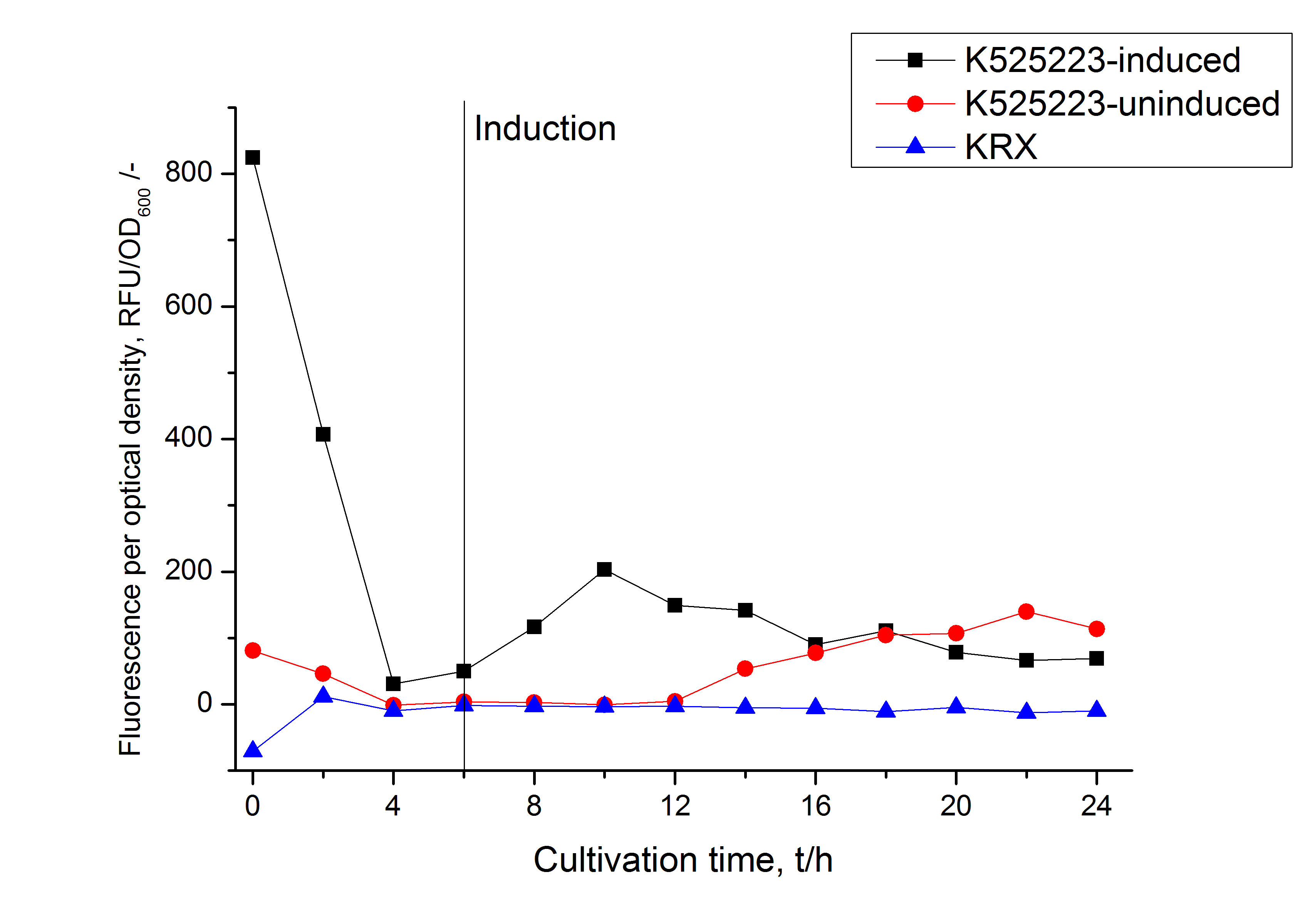
Identification and localisation
After a cultivation time of 18 h the mRFP|CspB fusion protein was localized in E. coli KRX. Therefore a part of the produced biomass was mechanically disrupted and the resulting lysate was washed with ddH2O. From the other part the periplasm was detached by using an osmotic shock.
The S-layer fusion protein could not be found in the polyacrylamide gel after a SDS-PAGE of the lysate. This indicated that the fusion protein integrates into the cell membrane with its lipid anchor. For testing this assumption the washed lysate was treated with ionic, nonionic and zwitterionic detergents to release the mRFP|CspB out of the membranes.
The existance of flourescence in two of the detergent fractions (10 % SDS and 10 % N-lauroyl sarcosine) and the low fluorescence in the wash fraction confirm the hypothesis of an insertion into the cell membrane (fig. 3). An insertion of these S-layer proteins might stabilize the membrane structure and increase the stability of cells against mechanical and chemical treatment. A stabilization of E. coli expressing S-layer proteins was described by [http://mic.sgmjournals.org/content/156/12/3584.long Lederer et al., 2010].
An other important fact is, that there is actually mRFP fluorescence measurable in such high concentrated detergent solutions. The S-layer seems to stabilize the biologically active conformation of mRFP.
In comparison with the mRFP fusion protein of K525224, wich has a TAT-sequence, a minor relative fluorescence in all cultivation and detergent fractions were detected (fig. 3). Together with the decreasing RFU after 9 h of cultivation (fig. 2) this results indicate a postive effect of the TAT-sequence on the protein stability. This could be due to a digestion of K525223 by proteases in the cytoplasm.
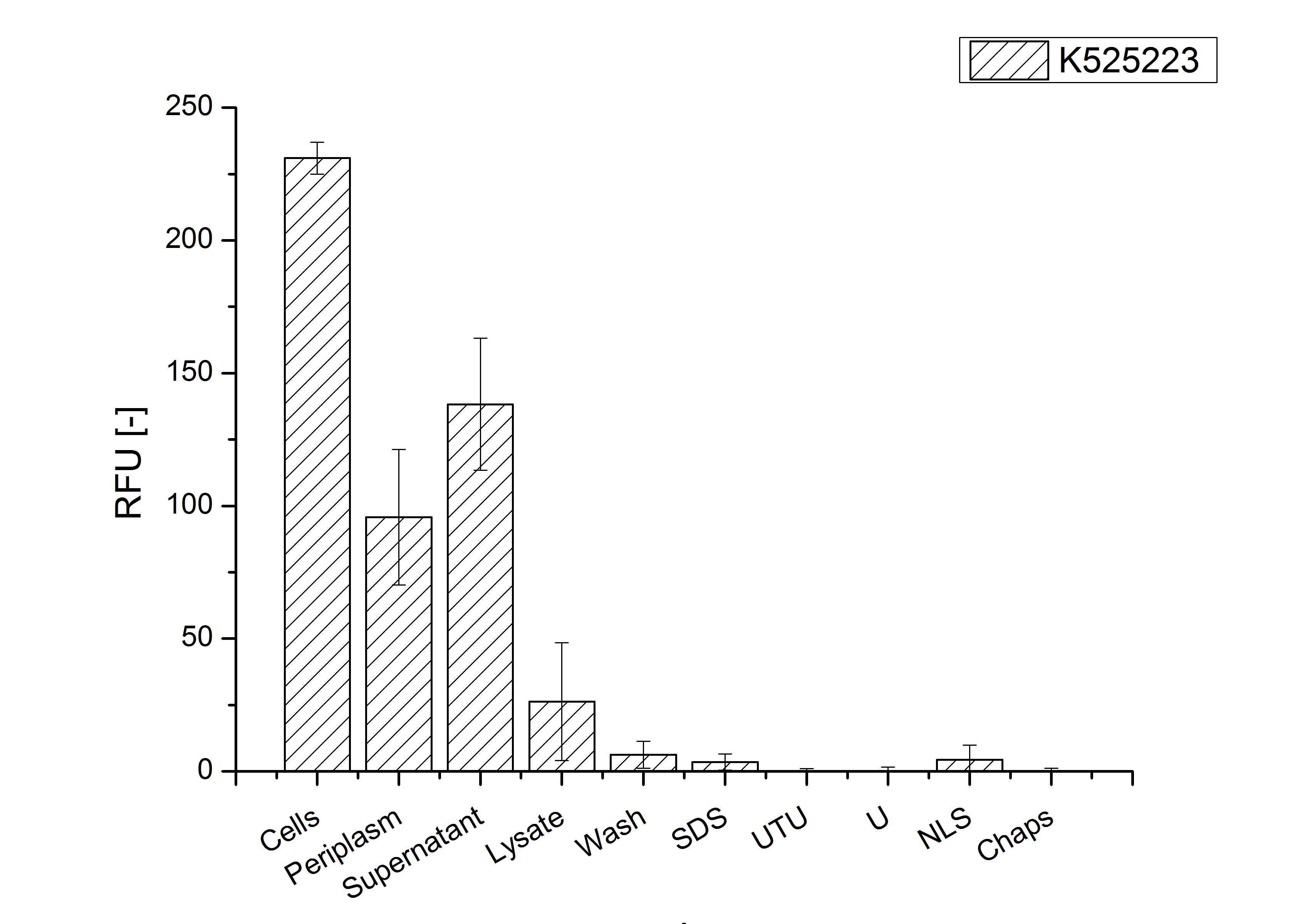
To obtain more specific information about the location of the S-layer fusion protein, after comparison with same treated fraction of E. coli KRX all gel bands in a defined size area were cut out of the gel and analysed with MALDI-TOF. Results are shown in fig. 4. The fusion protein CspB|mRFP (BBa_E1010) features a lipid anchor at the carboxy-terminus, but no amino-terminal TAT-sequence. In accordance with other protein variants with and without this features, the protein should be located mainly in the cytoplasm as inclusion bodies or incooperated with its lipid anchor into the cell membrane. Thus, the fraction with 10 % (v/v) SDS as detergent to disintegrate the protein from the cell wall was measured with MALDI-TOF. Results are shown in fig. 4.
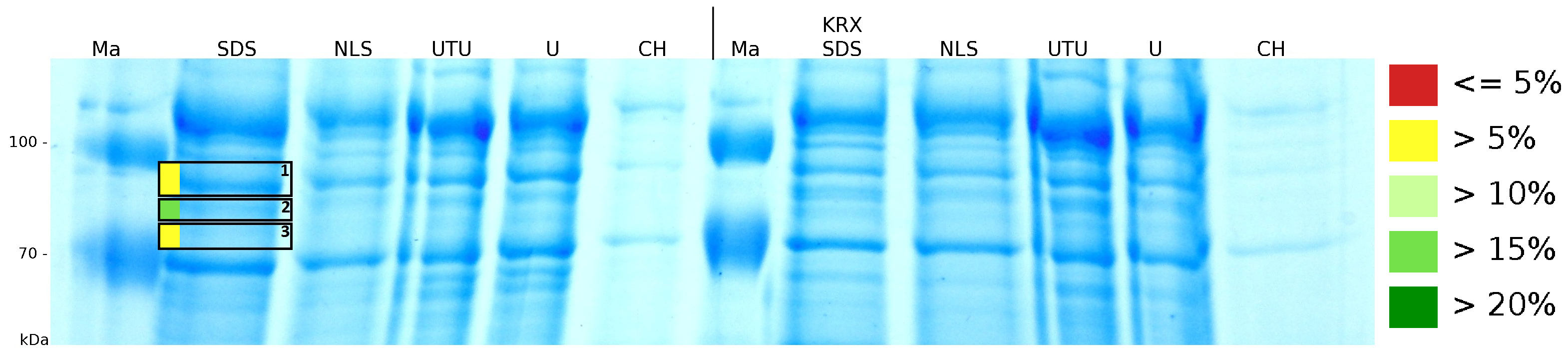
Fig. 4 shows, that the protein could be identified in all measured gel bands. The results indicate, that the protein is incorperated into the cell membrane. No fluoresence could be detected in the fractions using urea as detergent (see fig. 3), thus the protein probably does not form inclusion bodies. The fact that there is a comparable low overall fluorescence in the cells is a strong indication that the protein is degraded by E. coli proteases. The fluorescence in the periplasm and the supernatant is probably due to cell lysis during the periplasm isolation and the cultivation because the measured fluorescence values are comparable low, too.
Methods
Expression of S-layer genes in E. coli
- Chassis: Promega's [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX]
- Medium: LB medium supplemented with 20 mg L-1 chloramphenicol
- For autoinduction: Cultivations in LB-medium were supplemented with 0.1 % L-rhamnose as inducer and 0.05 % glucose
Measuring of mRFP
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 584 nm
- Emission: 620 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
Tryptic digest of gel lanes for analysis with MALDI-TOF
Note:
- Make sure to work under a fume hood.
- Do not work with protective gloves to prevent contamination of your sample with platicizers.
Reaction tubes have to be cleaned with 60% (v/v) CH3CN, 0.1% (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing.
- Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices.
- Gel slices should be washed two times. Therefore add 200 µL 30% (v/v) acetonitrile in 0.1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste.
- Dry gel slices at least 30 minutes in a Speedvac.
- Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation.
- Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night.
- Dry gel slices at least 30 minutes in a Speedvac.
- According to the size of the gel slice, add 5 – 20 µL 50% (v/v) ACN / 0,1% (v/v) TFA.
- Samples can be used for MALDI measurement or stored at -20 °C.
Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH4HCO3
- Therefore solubilize lyophilized Trypsin in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C.
Preparation and Spotting for analysis of peptides on Bruker AnchorChips
- Spot 0,5 – 1 µL sample aliquot
- Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip.
Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area.
- Dry the prepared spots at room temperature
- Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0,1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial.
- Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots.
References
Lederer FL, Günther TJ, Flemming K, Raff J, Fahmy K, Springer A, Pollmann K (2010) Heterologous expression of the surface-layer-like protein SllB induces the formation of long filaments of Escherichia coli consisting of protein-stabilized outer membrane. [http://mic.sgmjournals.org/content/156/12/3584.long Microbiology. 156(Pt 12):3584-95].
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].