Difference between revisions of "Part:BBa K3351002"
(→CONTRIBUTION: UTokyo 2021) |
(→Expression of recombinant antimicrobial peptide in E. coli) |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 9: | Line 9: | ||
<html> | <html> | ||
<img style="display: block; | <img style="display: block; | ||
| − | width: 60%;height: 60%;" src="https://static.igem.org/mediawiki/parts/d/d7/T--NWU-CHINA-A--od-2.png"><div>Figure.1 growth curve</div></html> | + | width: 60%;height: 60%;" src="https://static.igem.org/mediawiki/parts/d/d7/T--NWU-CHINA-A--od-2.png" text-align="center"><div>Figure.1 growth curve</div></html> |
===Reference=== | ===Reference=== | ||
| Line 50: | Line 50: | ||
HBD3 has antimicrobial effects on yeast not only on bacteria, so we evaluate the effect of HBD3 on <i>Saccharomyces cerevisiae</i>. | HBD3 has antimicrobial effects on yeast not only on bacteria, so we evaluate the effect of HBD3 on <i>Saccharomyces cerevisiae</i>. | ||
| − | + | ===Purpose=== | |
| − | + | The UTokyo 2021 aimed to make HBD3 secreting <i>S. cerevisiae</i> for wound care. However, it is reported that HBD3 has antimicrobial activity against yeast [1], and secretion of HBD3 may affect the growth of yeast. Therefore, the growth rate between the HBD3-secreting strain and the control strain had to be compared in our project. | |
| − | The UTokyo 2021 aimed to make HBD3 secreting <i>S. cerevisiae</i> for wound care. However, it is reported that HBD3 has antimicrobial activity against yeast [ | + | |
| − | + | ===Method=== | |
| − | + | To evaluate the effect of HBD3 on <i>S. cerevisiae</i>, we evaluated the growth rate of HBD3-secreting yeast according to the following protocol. HBD3 secreting yeast was made by transferring ( <partinfo>BBa_K3804013</partinfo>) | |
| − | To evaluate the effect of HBD3 on <i>S. cerevisiae</i>, we evaluated the growth rate of HBD3-secreting yeast according to the following protocol. HBD3 secreting yeast was made by transferring ( | + | |
<br>1. Inoculate 200ul of saturated yeast culture into 3ml of YPD. | <br>1. Inoculate 200ul of saturated yeast culture into 3ml of YPD. | ||
<br>3. Incubate at 30°C and 200 rpm with shaking. | <br>3. Incubate at 30°C and 200 rpm with shaking. | ||
| − | <br>2. Measure the | + | <br>2. Measure the OD600 of the culture medium at regular intervals. |
| − | As a control, we use pRS316(which is a shuttle vector for yeast and <i>E. coli</i>) transferred | + | As a control, we use pRS316(which is a shuttle vector for yeast and <i>E. coli</i>) transferred <i>S. cerevisiae</i>. |
| − | < | + | ===Results=== |
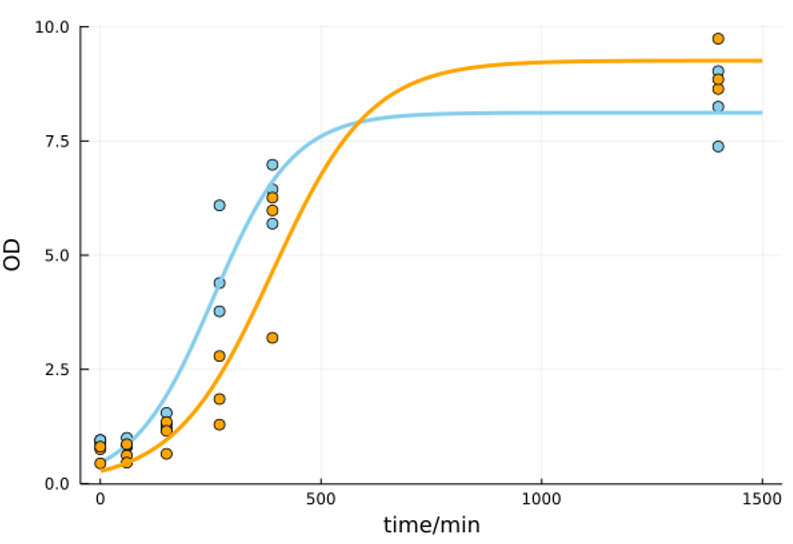
| + | As shown in Figure 2, HBD3 affected yeast growth during the exponential phase compared to control. This property was observed particularly at 250 minutes after the inoculation. However, since the final concentration was almost the same, HBD3 secretion was considered to have little effect on the number of <i>S. cerevisiae</i> in the end. Thus, HBD3 secretion yeast is expected to be sufficient for practical implementation. | ||
| + | [[File:T--UTokyo--HBD3_Yeast.png|550px|thumb|center|Figure2. Effect of HBD3 expression on growth of <i>S. cerevisiae</i>. Orange:HBD3 expressing strain. Blue:control strain.]]<br> | ||
| + | |||
| + | ===Reference=== | ||
| + | [1] J. Harder, J. Bartels, E. Christophers, and J.-M. Schröder, “Isolation and Characterization of Human μ-Defensin-3, a Novel Human Inducible Peptide Antibiotic,” J. Biol. Chem., vol. 276, no. 8, pp. 5707–5713, Feb. 2001, doi: 10.1074/jbc.M008557200. | ||
<br> | <br> | ||
| − | + | ||
| + | ==CONTRIBUTION: LINKS-China 2024 == | ||
| + | Authors iGEM LINKS-China 2024, Zhengyu He. | ||
| + | ===Expression of recombinant antimicrobial peptide in E. coli=== | ||
| + | This year we used SHuffle T7 to express HBD3 with a 6×His tag, aiming to purify and test its antimicrobial activity. However, we did not observe the correct target protein (6.4kDa) on Tricine-SDS-PAGE, and the growth of the expression strain was not inhibited. This may be due to the small molecular weight leading to rapid proteolytic degradation. | ||
| + | |||
<html> | <html> | ||
| − | + | <img style="display: block; | |
| − | + | width: 60%;height: 60%;" src="https://static.igem.wiki/teams/5185/part-org/hbd3/hbd3-1.jpg" text-align="center"><div>Figure.1 SDS-PAGE analysis of the recombinant defensins expression in E. coli SHuffle T7</div></html> | |
| − | </html> | + | |
| + | ===Expression of recombinant antimicrobial peptide in E. coli=== | ||
| + | To successfully produce HBD3, we added a CBM3-SUMO tag at the N-terminus of HBD3, and then used Ulp1 enzyme for cleavage to release HBD3. | ||
| + | <html> | ||
| + | <img style="display: block; | ||
| + | width: 60%;height: 60%;" src="https://static.igem.wiki/teams/5185/part-org/hbd3/hbd3-3.jpg" text-align="center"><div>Figure 2: Characterization of fused HBD3 in (A) Induction expression of CBM3-SUMO-HBD3. (B) Sumo enzyme cleavage validation of CBM3-SUMO-HBD3. (C) Antibacterial test of CBM3-SUMO↓HBD3. (D) Impact of different concentrations of CBM3-SUMO↓HBD3 on the growth of Staphylococcus aureus. (E) Antibacterial ability of different concentrations of CBM3-SUMO↓HBD3 after 8 hours.</div></html> | ||
| + | |||
| + | 4 types of CBM3-SUMO-Defensins were subjected to salt removal by gradient dialysis, followed by cleavage with recombinant Ulp1 (Beyotime, P2312S). The results from SDS-PAGE electrophoresis showed a slight decrease in the molecular weight of the target protein (Fig. 2 B), indicating successful removal of ~4 kDa defensins. Since CBM3-SUMO-Defensins would ultimately be incorporated into wound dressing products in a domain-bound form rather than as individual defensins, we did not further purify the defensins. Instead, we utilized the enzyme-cleaved CBM3-SUMO-Defensins (designated as CBM3-SUMO↓Defensins) for antimicrobial assays. As depicted in Fig. 2C, Escherichia coli and Staphylococcus aureus were selected as representatives of Gram-positive and Gram-negative bacteria, respectively. The CBM3-SUMO↓Defensins cleaved by Ulp1 enzyme exhibited antimicrobial activity against both strains, while the uncleaved CBM3-SUMO-Defensins showed no antimicrobial activity. This suggests that we successfully produced active defensin molecules using the fusion protein cleavage approach. | ||
| + | |||
| + | we utilized the microdilution method to determine the MIC values of four types of CBM3-SUMO-Defensins. For specific details, please refer to our measurement section. Initially, we examined the 24-hour growth curves of Staphylococcus aureus with the addition of CBM3-SUMO↓Defensins. Within the 0–8 hour range, all four types of CBM3-SUMO↓Defensins exhibited antimicrobial activity (Fig. 2E and 2F). We selected the 8-hour time point to define the MIC values against Staphylococcus aureus. At this point, the MIC50 values for CBM3-SUMO↓HNP1, CBM3-SUMO↓HNP4, CBM3-SUMO↓HD5, and CBM3-SUMO↓HBD3 were 0.74 μM, 0.368 μM, 1.475 μM, and 1.001 μM, respectively. Additionally, the MIC90 values for CBM3-SUMO↓HNP4/HD5 were 0.735 μM and 1.475 μM, respectively. These values are close to the MIC values reported in the literature for the four defensins[1]. | ||
| + | |||
| + | ===Reference=== | ||
| + | Wei, G., de Leeuw, E., Pazgier, M., Yuan, W., Zou, G., Wang, J., ... & Lu, W. (2009). Through the looking glass, mechanistic insights from enantiomeric human defensins. Journal of Biological Chemistry, 284(42), 29180-29192. | ||
Latest revision as of 13:24, 1 October 2024
HBD3, an antimicrobial peptide.
Summary
The three human β-defensins, HBD1-3, are 33-47-residue, cationic antimicrobial proteins expressed by epithelial cells. All three proteins have broad spectrum antimicrobial activity, with HBD3 consistently being the most potent. Additionally, HBD3 has significant bactericidal activity against Gram-positive Staphylococcus aureus at physiological salt concentrations.Defensins are small, 3–5 kDa cationic proteins constrained by three disulfide bonds. As a class of proteins, they have broad microbicidal activity against Gram-positive and -negative bacteria, yeast, and some enveloped viruses, although specific defensin peptides often have defined spectra of activity. Like many other antimicrobial peptides, the defensin class of peptides is known to disrupt the membranes of microbes . It has recently been reported that in addition to their antimicrobial activity, defensins may act as chemokines, activating the adaptive immune response .HBD3 possesses bactericidal activity against Gram-positive and -negative bacteria, including multi-drug-resistant S. aureus, vancomycin-resistant Enterococcus faecium, and Burkholderia cepacia in addition to the yeast C. albicans .
Characterization

Reference
[1] David J,Schibli,Howard N,Hunter,Vladimir,Aseyev,Timothy D,Starner,John M,Wiencek,Paul B,McCray,Brian F,Tack,Hans J,Vogel.The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.[J].The Journal of biological chemistry,2002,277(10):8279-89.
[2] Schroder, J. M. (1999) Cell. Mol. Life Sci. 56, 32–46
[3] Epand, R. M., and Vogel, H. J. (1999) Biochim. Biophys. Acta 1462, 11–28
[4] Harder, J., Bartels, J., Christophers, E., and Schroder, J. M. (2001) J. Biol. Chem. 276, 5707–5713
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
CONTRIBUTION: UTokyo 2021
Authors iGEM UTokyo 2021, Rintaro Shimojo.
HBD3 has antimicrobial effects on yeast not only on bacteria, so we evaluate the effect of HBD3 on Saccharomyces cerevisiae.
Purpose
The UTokyo 2021 aimed to make HBD3 secreting S. cerevisiae for wound care. However, it is reported that HBD3 has antimicrobial activity against yeast [1], and secretion of HBD3 may affect the growth of yeast. Therefore, the growth rate between the HBD3-secreting strain and the control strain had to be compared in our project.
Method
To evaluate the effect of HBD3 on S. cerevisiae, we evaluated the growth rate of HBD3-secreting yeast according to the following protocol. HBD3 secreting yeast was made by transferring ( BBa_K3804013)
1. Inoculate 200ul of saturated yeast culture into 3ml of YPD.
3. Incubate at 30°C and 200 rpm with shaking.
2. Measure the OD600 of the culture medium at regular intervals.
As a control, we use pRS316(which is a shuttle vector for yeast and E. coli) transferred S. cerevisiae.
Results
As shown in Figure 2, HBD3 affected yeast growth during the exponential phase compared to control. This property was observed particularly at 250 minutes after the inoculation. However, since the final concentration was almost the same, HBD3 secretion was considered to have little effect on the number of S. cerevisiae in the end. Thus, HBD3 secretion yeast is expected to be sufficient for practical implementation.
Reference
[1] J. Harder, J. Bartels, E. Christophers, and J.-M. Schröder, “Isolation and Characterization of Human μ-Defensin-3, a Novel Human Inducible Peptide Antibiotic,” J. Biol. Chem., vol. 276, no. 8, pp. 5707–5713, Feb. 2001, doi: 10.1074/jbc.M008557200.
CONTRIBUTION: LINKS-China 2024
Authors iGEM LINKS-China 2024, Zhengyu He.
Expression of recombinant antimicrobial peptide in E. coli
This year we used SHuffle T7 to express HBD3 with a 6×His tag, aiming to purify and test its antimicrobial activity. However, we did not observe the correct target protein (6.4kDa) on Tricine-SDS-PAGE, and the growth of the expression strain was not inhibited. This may be due to the small molecular weight leading to rapid proteolytic degradation.

Expression of recombinant antimicrobial peptide in E. coli
To successfully produce HBD3, we added a CBM3-SUMO tag at the N-terminus of HBD3, and then used Ulp1 enzyme for cleavage to release HBD3.

4 types of CBM3-SUMO-Defensins were subjected to salt removal by gradient dialysis, followed by cleavage with recombinant Ulp1 (Beyotime, P2312S). The results from SDS-PAGE electrophoresis showed a slight decrease in the molecular weight of the target protein (Fig. 2 B), indicating successful removal of ~4 kDa defensins. Since CBM3-SUMO-Defensins would ultimately be incorporated into wound dressing products in a domain-bound form rather than as individual defensins, we did not further purify the defensins. Instead, we utilized the enzyme-cleaved CBM3-SUMO-Defensins (designated as CBM3-SUMO↓Defensins) for antimicrobial assays. As depicted in Fig. 2C, Escherichia coli and Staphylococcus aureus were selected as representatives of Gram-positive and Gram-negative bacteria, respectively. The CBM3-SUMO↓Defensins cleaved by Ulp1 enzyme exhibited antimicrobial activity against both strains, while the uncleaved CBM3-SUMO-Defensins showed no antimicrobial activity. This suggests that we successfully produced active defensin molecules using the fusion protein cleavage approach.
we utilized the microdilution method to determine the MIC values of four types of CBM3-SUMO-Defensins. For specific details, please refer to our measurement section. Initially, we examined the 24-hour growth curves of Staphylococcus aureus with the addition of CBM3-SUMO↓Defensins. Within the 0–8 hour range, all four types of CBM3-SUMO↓Defensins exhibited antimicrobial activity (Fig. 2E and 2F). We selected the 8-hour time point to define the MIC values against Staphylococcus aureus. At this point, the MIC50 values for CBM3-SUMO↓HNP1, CBM3-SUMO↓HNP4, CBM3-SUMO↓HD5, and CBM3-SUMO↓HBD3 were 0.74 μM, 0.368 μM, 1.475 μM, and 1.001 μM, respectively. Additionally, the MIC90 values for CBM3-SUMO↓HNP4/HD5 were 0.735 μM and 1.475 μM, respectively. These values are close to the MIC values reported in the literature for the four defensins[1].
Reference
Wei, G., de Leeuw, E., Pazgier, M., Yuan, W., Zou, G., Wang, J., ... & Lu, W. (2009). Through the looking glass, mechanistic insights from enantiomeric human defensins. Journal of Biological Chemistry, 284(42), 29180-29192.