Difference between revisions of "Part:BBa K2151200"
| (10 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2151200 short</partinfo> | <partinfo>BBa_K2151200 short</partinfo> | ||
| − | |||
This biobrick was created through standard biobrick assembly of K118014(RBS+crtE), K118006(RBS+crtB), K118005(RBS+crtI) and K118013(crtY). These genes are a part of the carotenoid biosynthesis pathway and together, this biobrick converts converts colourless farnesyl pyrophosphate to orange beta-carotene | This biobrick was created through standard biobrick assembly of K118014(RBS+crtE), K118006(RBS+crtB), K118005(RBS+crtI) and K118013(crtY). These genes are a part of the carotenoid biosynthesis pathway and together, this biobrick converts converts colourless farnesyl pyrophosphate to orange beta-carotene | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 14: | Line 13: | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| − | |||
<partinfo>BBa_K2151200 parameters</partinfo> | <partinfo>BBa_K2151200 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| − | + | ||
| − | As many researches indicate, the major problem of polycistronic vectors, which contain two or more target genes under one promoter, is the much lower expression of the downstream genes compared with that of the first gene next to the promoter<ref>Kim, K. J., Kim, H. E., Lee, K. H., Han, W., Yi, M. J., Jeong, J., & Oh, B. H. (2004). Two-promoter vector is highly efficient for overproduction of protein complexes. Protein science : a publication of the Protein Society, 13(6), 1698–1703.</ref>. Instead of assembling CDSs sequentially, we construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between crtEBIY. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons ''in vivo''. Self-interaction of the polycistron can be | + | |
| + | ===Note added by Team Fudan 2022=== | ||
| + | |||
| + | As many researches indicate, the major problem of polycistronic vectors, which contain two or more target genes under one promoter, is the much lower expression of the downstream genes compared with that of the first gene next to the promoter<ref>Kim, K. J., Kim, H. E., Lee, K. H., Han, W., Yi, M. J., Jeong, J., & Oh, B. H. (2004). Two-promoter vector is highly efficient for overproduction of protein complexes. Protein science : a publication of the Protein Society, 13(6), 1698–1703.</ref>. Even though the original design here put crt genes order as how β-carotene was produced (crtE → crtB → crtI → crtY), the team failed to present data supporting their design works. | ||
| + | |||
| + | Instead of assembling CDSs sequentially, we construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between crtEBIY. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons ''in vivo''. Self-interaction of the polycistron can be avoid and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons, as in [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117]. | ||
| + | |||
| + | ====Improved part by Team Fudan 2022==== | ||
| + | Our improved part is [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117] and [https://parts.igem.org/Part:BBa_K4162118 BBa_K4162118]. We not only using ribozyme sequence flanking '''RBS+CDS''', but also using [https://parts.igem.org/Part:BBa_B0030 a stronger RBS] to drive crtE expression in biobrick crtEBIY. | ||
| + | |||
| + | ====Successful production of β-carotene in bacteria DH5α==== | ||
| + | Our [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117] biobrick was created through overlapping PCR of [https://parts.igem.org/Part:BBa_K4162020 BBa_K4162020](ribozyme+J6_RBS+crtY), [https://parts.igem.org/Part:BBa_K4162010 BBa_K4162010](ribozyme+T7_RBS+crtE), [https://parts.igem.org/Part:BBa_K4162013 BBa_K4162013](ribozyme+T7_RBS+crtB) and [https://parts.igem.org/Part:BBa_K4162016 BBa_K4162016](ribozyme+T7_RBS+crtI). We transfected [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117] into ''E. coli'' DH5α to build single-cell factory for β-carotene production. Coding sequences of crtYEBI are separated by ribozyme sequences. In this part, the RBS of crtEBI has equal intensity while the RBS of crtY is significantly weaker than the others. Because crtY catalyzes the last step of the carotenoid reaction chain, we guess the concentration of substrate catalyzed by this enzyme is significantly lower than for the first three steps of the reaction. To avoid the problem of flux imbalance in biosynthesis as well as to reduce unnecessary metabolic stress on cells, we intentionally weakened the RBS intensity of crtY, and created [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117]. Please notice, our [https://parts.igem.org/Part:BBa_K4162118 BBa_K4162118] places crt genes in the same order as [https://parts.igem.org/Part:BBa_K2151200 BBa_K2151200], but not [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117]. | ||
| + | |||
| + | ===Characterization of crtYEBI=== | ||
| + | ====Agarose gel electrophoresis==== | ||
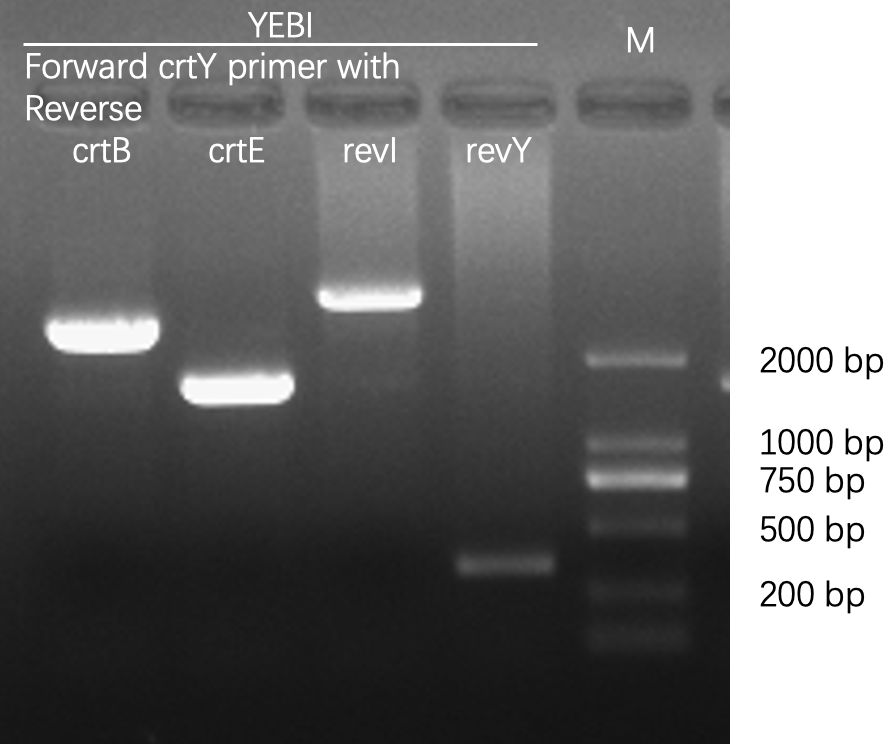
| + | [[File:T--Fudan--Agarose gel electrophoresis--crtYEBI.png|400px|thumb|none|'''Figure 1. Agarose gel electrophoresis of PCR products, amplified from bacterial colonies/cultures.''' The first lane was loaded with D2000 DNA ladder whose sizes were marked on the image. We chose Taq DNA polymerase for its low cost and high reliability, and we designed forward and reverse primers for each carotene synthesis enzyme (crt for short). The PCR reaction was composed of 2 μL 10x Taq polymerase buffer, 16 μL H2O, 0.5 μL Taq polymerase, 0.5 μL dNTP (10 mM each), 0.5 μL forward primer (10 mM), 0.5 μL reverse primer (10 mM), and 1 μL bacterial culture or 1μL colony. Using the same forward primer, and different reverse primers, we were able to detect the composition of various crt genes. After PCR, the correct bacterial clones were sent for Sanger sequencing. Once verified, these clones would be used for further experiments. The sequences of primers are: > 5-crtY 5-ATGCAACCGCATTATGATCTGATTC-3; > rev320crtB 5-CCTTCCAGATGATCAAACGCGTAAG-3; > rev320crtE 5-ATGAGAATGAATGGTAGGGCGTC-3; > rev320crtI 5-GGATTAAACTGCTGAATCTGCGCTTC-3; > rev320crtY 5-CCGCGGTATCCATCCACAAG-3.]] | ||
| + | |||
| + | ====Successful protein expression==== | ||
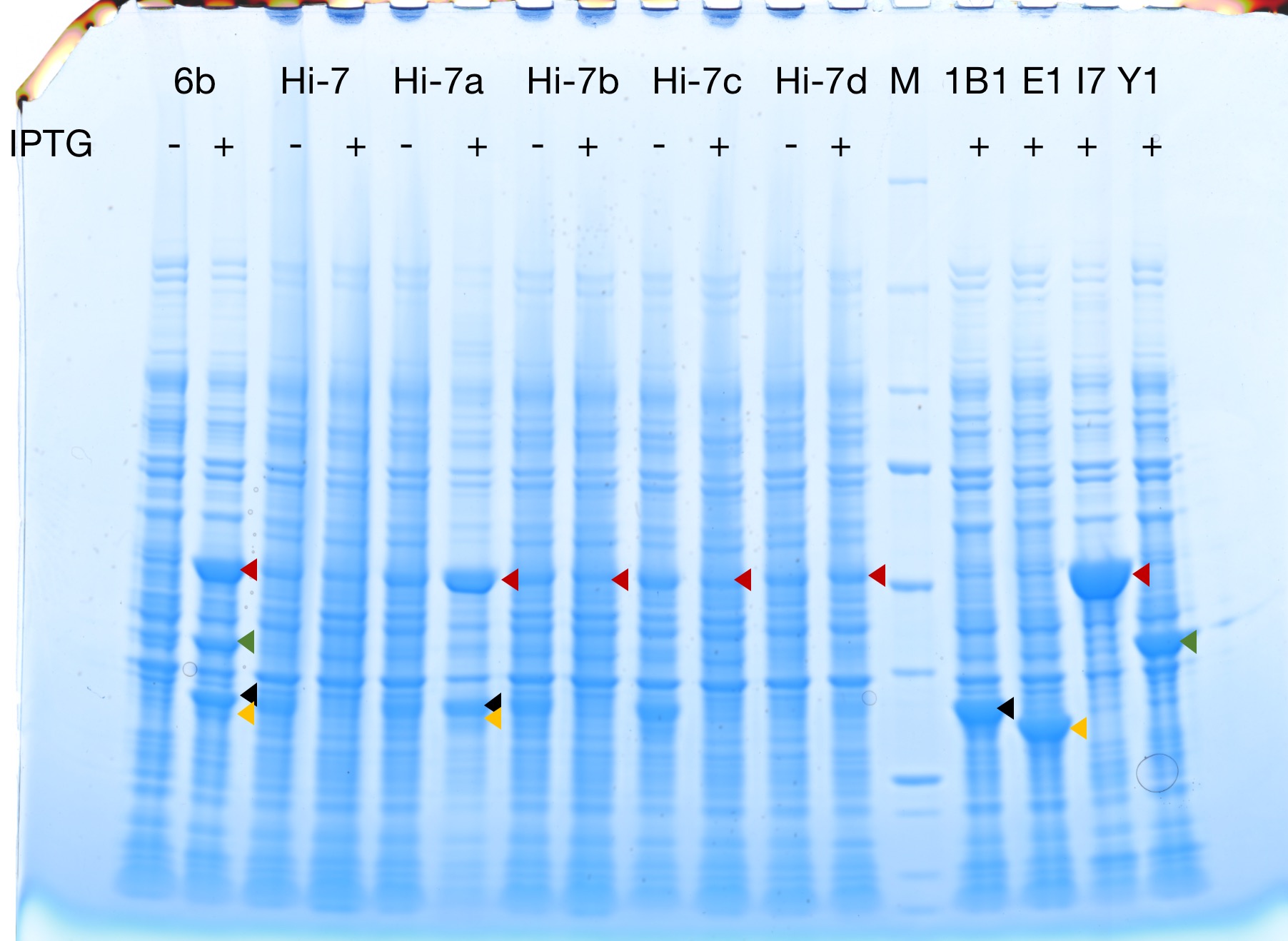
| + | [[File:T--Fudan--Hi--117.png|400px|thumb|none|'''Figure 2. SDS-PAGE.'''IPTG(-/+) = without/with 0.2 mM IPTG for 3-6 hours, adding IPTG to a bacteria culture with OD600 0.2-0.3. M: Protein molecular weight marker ladder. Lane 1~2: pET28 plasmids encoding crtEBIY without any tag were transformed into BL21(DE3) HI-Control strain, single clones (6b) were picked for liquid LB culture. Lane 3~12: pET28 plasmids encoding crtYEBI without any tag were transformed into BL21(DE3) Hi-Control strain, single clones (Hi-7, Hi-7a, Hi-7b, Hi-7c, Hi-7d) were picked for liquid LB culture. Lane 14~17: pET28 plasmids encoding crtB, crtE, crtI, crtYwithout any tag were transformed into BL21(DE3) HI-Control strain, single clones (1B1, E1, I7, Y1) were picked for liquid LB culture. Protein expression was induced in parallel cultures by IPTG. Bacterial cultures were monitored by OD600, and 5x10^7 cells were harvested by centrifugation and lysis in 1x SDS sample buffer. Equal amount (10 μL, 2x10^6 cells) of whole cell lysate were analyzed by SDS-PAGE (4~20% gradient gel, Tanon brand). Red arrows point to crtI protein. Green arrows point to crtY protein. Black arrows point to crtB protein. Yellow arrows point to crtE protein.]] | ||
| + | |||
| + | ====Produce β-carotene==== | ||
| + | Figures 2 to 5 show that ''E. coli'' transfected with [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117] successfully expressed the target enzyme and yielded β-carotene. In Figure 5, it can be seen that module YEBI corresponds to a darker orange color of the post-centrifugation precipitation compared to module YBEI ([https://parts.igem.org/Part:BBa_K4162119 BBa_K4162119]), characterizing the superior carotenoid yielding ability of module YEBI. | ||
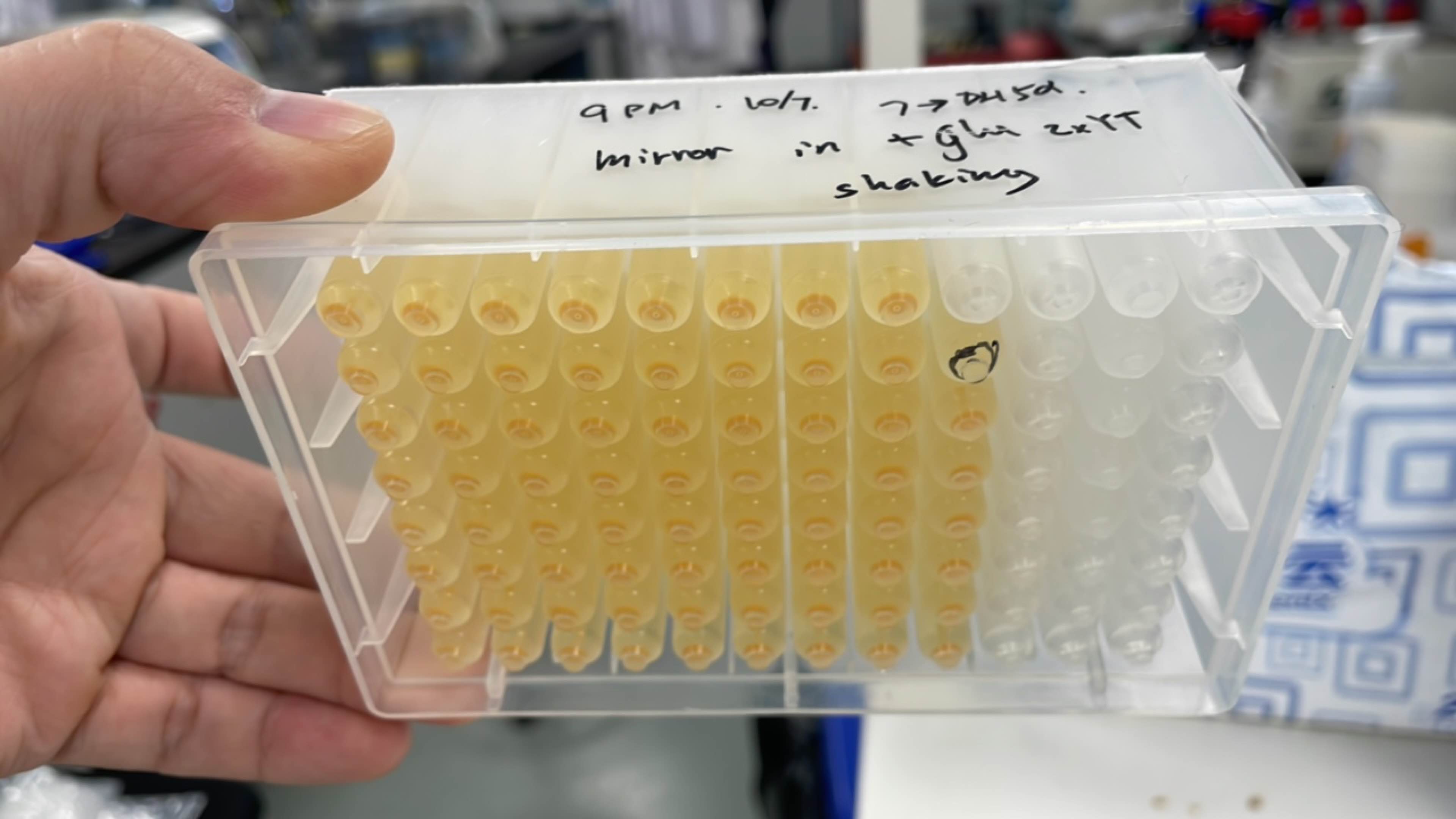
| + | [[File:T--Fudan--96a.png|400px|thumb|none|'''Figure 3. 96-well plate of module crtYEBI.''' Except for the blank control well marked in black, all clones growing different wells had similar β-carotene content in the bacterial pellet.]] | ||
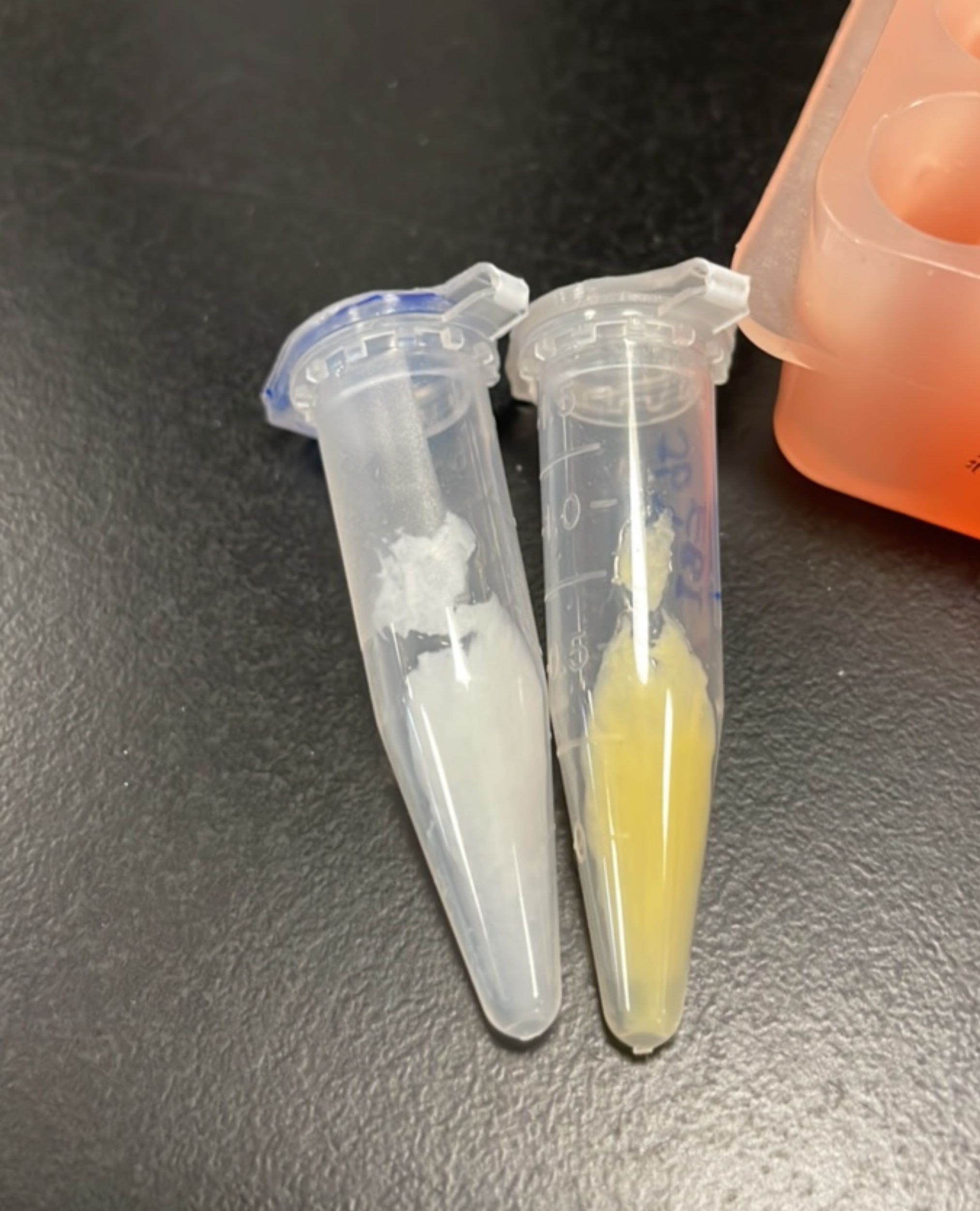
| + | [[File:T--Fudan--YEBI--117.png|400px|thumb|none|'''Figure 4.''' The centrifuge tube containing a visible yellow precipitation on the right is the module crtYEBI. The bacterial pellet was proceeded for [https://www.protocols.io/view/plasmid-miniprep-for-2022-dm6gpjwdpgzp/v1 miniprep]. After P1→P2→P3, 10-minute centrifugation and transfering the DNA containing supernatant into DNA binding column, we noticed what left, usually white cloudy precipitation, was yellow! Later, when we [https://www.protocols.io/view/acetone-extraction-of-bacteria-pellet-for-hplc-202-eq2ly7peelx9/v1 prepared samples for HPLC] from bacterial pellet, we show that these yellow ''stuff'' can be extracted into acetone.]] | ||
| + | [[File:T--Fudan--117--119.png|400px|thumb|none|'''Figure 5.''' The centrifuge tubes containing module crtYEBI (first from the left) and module crtYBEI [https://parts.igem.org/Part:BBa_K4162119 BBa_K4162119] (second from the left) contain visible yellow bacterial pellet.]] | ||
| + | |||
| + | ====HPLC validation==== | ||
| + | Agilent liquid chromatograph (HPLC-DAD); column C18 (250mm); column temperature 30°C; mobile phase methanol:water = 96:4; flow rate 0.8 mL/min; detection wavelength 325 nm and 454 nm. | ||
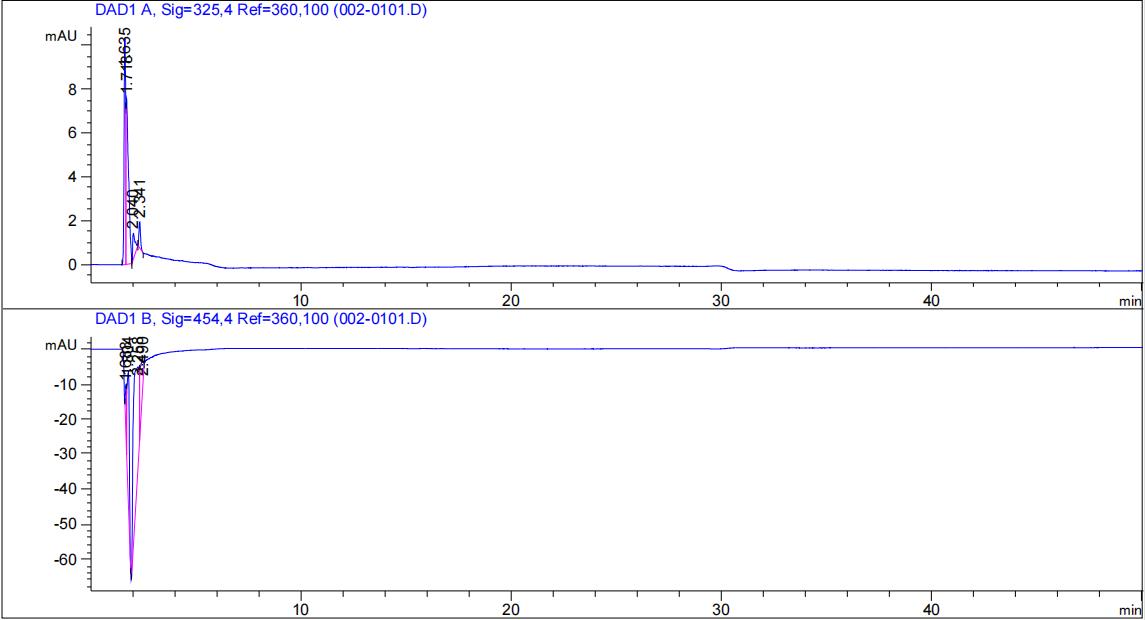
| + | [[File:T--Fudan--beta--unpuri--hplc.png|500px|thumb|none|'''Figure 6. Absorption peak of β-carotene standard solution. '''1:10 dilution of saturated solution of β-carotene in methanol.]] | ||
| + | [[File:T--Fudan--ecoli--unpuri--hplc.png|500px|thumb|none|'''Figure 7. Absorption peak of bacterial extracted sample. '''The β-carotene output of ''E. coli'' DH5α transfected with the plasmid carrying [https://parts.igem.org/Part:BBa_K4162117 BBa_K4162117] is shown in the graph has almost identical absorption peak pattern.]] | ||
<h2>'''References'''</h2> | <h2>'''References'''</h2> | ||
<references /> | <references /> | ||
Latest revision as of 05:35, 13 October 2022
crtEBIY
This biobrick was created through standard biobrick assembly of K118014(RBS+crtE), K118006(RBS+crtB), K118005(RBS+crtI) and K118013(crtY). These genes are a part of the carotenoid biosynthesis pathway and together, this biobrick converts converts colourless farnesyl pyrophosphate to orange beta-carotene Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1974
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1510
Illegal NgoMIV site found at 1640
Illegal AgeI site found at 725 - 1000COMPATIBLE WITH RFC[1000]
Note added by Team Fudan 2022
As many researches indicate, the major problem of polycistronic vectors, which contain two or more target genes under one promoter, is the much lower expression of the downstream genes compared with that of the first gene next to the promoter[1]. Even though the original design here put crt genes order as how β-carotene was produced (crtE → crtB → crtI → crtY), the team failed to present data supporting their design works.
Instead of assembling CDSs sequentially, we construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between crtEBIY. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Self-interaction of the polycistron can be avoid and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons, as in BBa_K4162117.
Improved part by Team Fudan 2022
Our improved part is BBa_K4162117 and BBa_K4162118. We not only using ribozyme sequence flanking RBS+CDS, but also using a stronger RBS to drive crtE expression in biobrick crtEBIY.
Successful production of β-carotene in bacteria DH5α
Our BBa_K4162117 biobrick was created through overlapping PCR of BBa_K4162020(ribozyme+J6_RBS+crtY), BBa_K4162010(ribozyme+T7_RBS+crtE), BBa_K4162013(ribozyme+T7_RBS+crtB) and BBa_K4162016(ribozyme+T7_RBS+crtI). We transfected BBa_K4162117 into E. coli DH5α to build single-cell factory for β-carotene production. Coding sequences of crtYEBI are separated by ribozyme sequences. In this part, the RBS of crtEBI has equal intensity while the RBS of crtY is significantly weaker than the others. Because crtY catalyzes the last step of the carotenoid reaction chain, we guess the concentration of substrate catalyzed by this enzyme is significantly lower than for the first three steps of the reaction. To avoid the problem of flux imbalance in biosynthesis as well as to reduce unnecessary metabolic stress on cells, we intentionally weakened the RBS intensity of crtY, and created BBa_K4162117. Please notice, our BBa_K4162118 places crt genes in the same order as BBa_K2151200, but not BBa_K4162117.
Characterization of crtYEBI
Agarose gel electrophoresis
Successful protein expression
Produce β-carotene
Figures 2 to 5 show that E. coli transfected with BBa_K4162117 successfully expressed the target enzyme and yielded β-carotene. In Figure 5, it can be seen that module YEBI corresponds to a darker orange color of the post-centrifugation precipitation compared to module YBEI (BBa_K4162119), characterizing the superior carotenoid yielding ability of module YEBI.

HPLC validation
Agilent liquid chromatograph (HPLC-DAD); column C18 (250mm); column temperature 30°C; mobile phase methanol:water = 96:4; flow rate 0.8 mL/min; detection wavelength 325 nm and 454 nm.

References
- ↑ Kim, K. J., Kim, H. E., Lee, K. H., Han, W., Yi, M. J., Jeong, J., & Oh, B. H. (2004). Two-promoter vector is highly efficient for overproduction of protein complexes. Protein science : a publication of the Protein Society, 13(6), 1698–1703.