Difference between revisions of "Part:BBa K4115040"
(→Results) |
|||
| (17 intermediate revisions by the same user not shown) | |||
| Line 21: | Line 21: | ||
|} | |} | ||
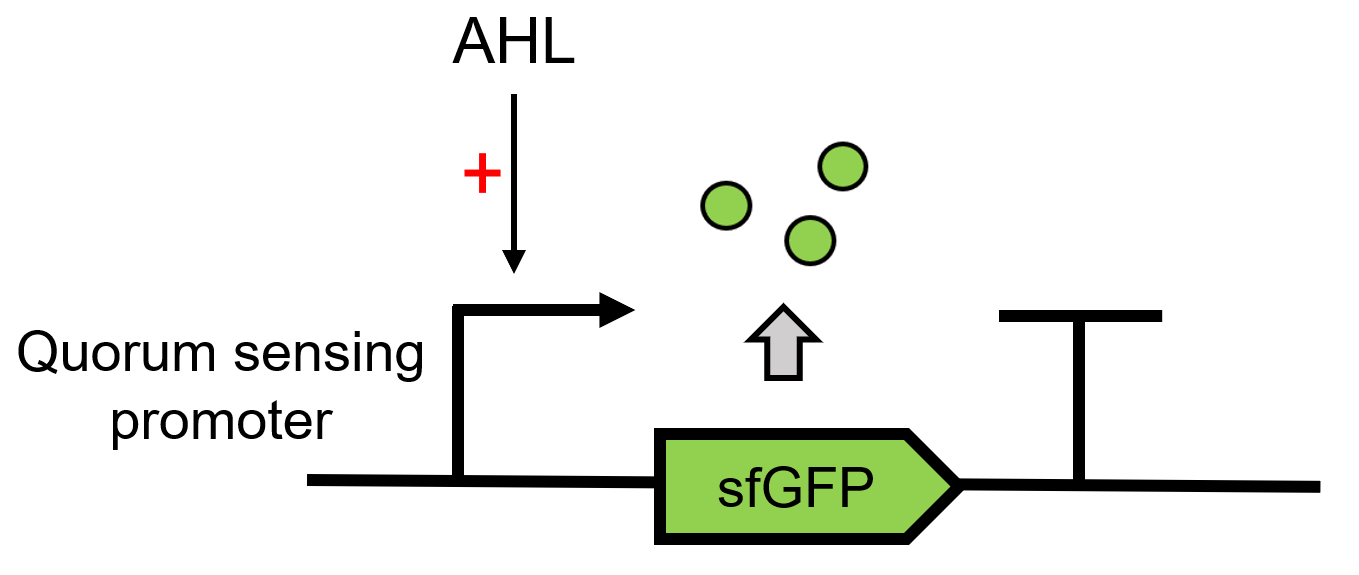
| − | This composite part is used to produce 3OC6 HSL by LacI [https://parts.igem.org/Part:BBa_C0161 (BBa_C0161)][1] under a constitutive promoter J23101[https://parts.igem.org/Part:BBa_J23101 (BBa_J23101)] and send it to the receiver protein LuxR [https://parts.igem.org/Part:BBa_C0062 (BBa_C0062)]in the other <i>E. coli</i>.When the LuxR receives the signal, it will regulate and express the downstream report gene sfGFP [2], | + | This composite part is used to produce 3OC6 HSL by LacI [https://parts.igem.org/Part:BBa_C0161 (BBa_C0161)][1] under a constitutive promoter J23101[https://parts.igem.org/Part:BBa_J23101 (BBa_J23101)] and send it to the receiver protein LuxR [https://parts.igem.org/Part:BBa_C0062 (BBa_C0062)]in the other <i>E. coli</i>.When the LuxR receives the signal, it will regulate and express the downstream report gene sfGFP [2], Figure 1. Thus we can detect whether LuxI is functional by observing the fluorescence intensity under UV light. |
| + | |||
| + | [[File:Results-QS report.png|550px|thumb|left|'''Figure 1:''' Lux QS system path.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
==Proof of function== | ==Proof of function== | ||
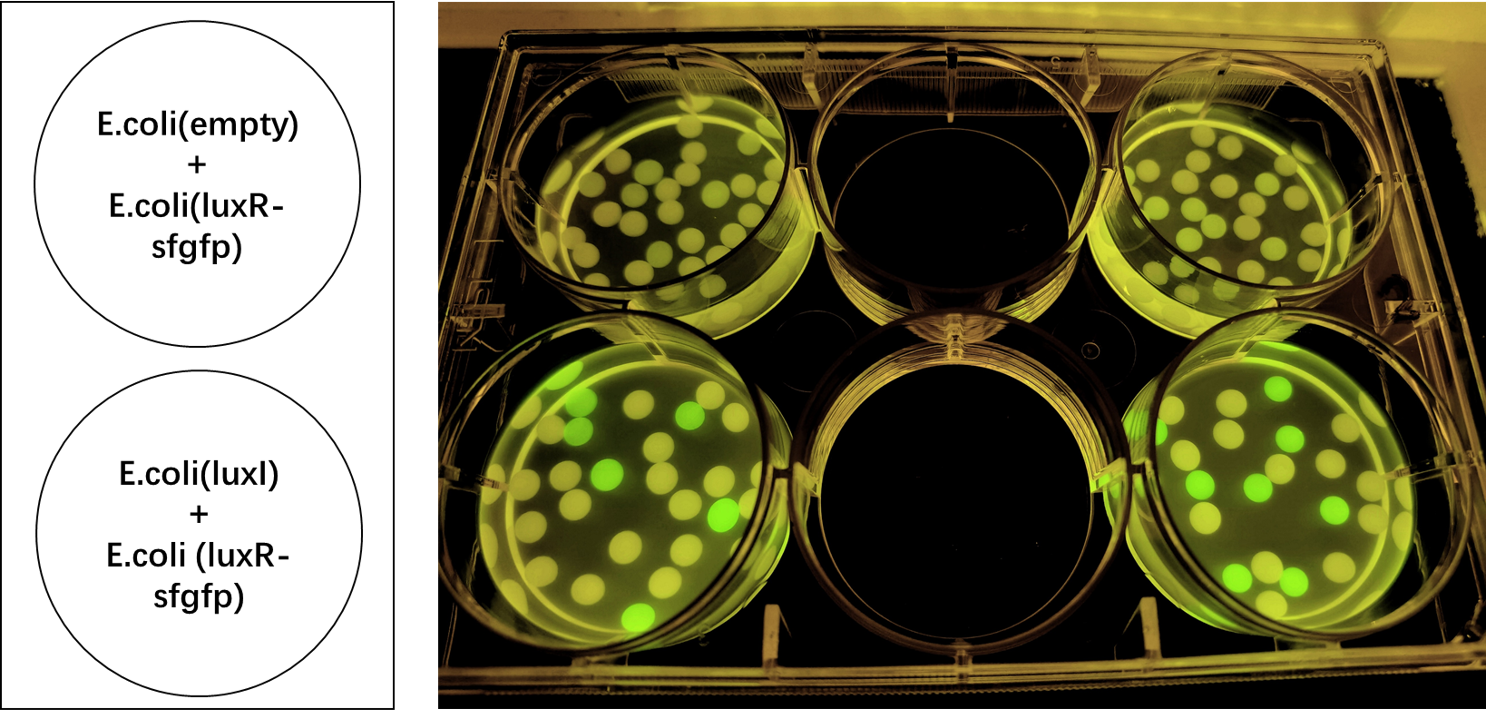
| − | We encapsulated <i>E. coli</i> with this composite part and <i>E. coli</i> with LuxR and sfGFP into alginate gel beads. The <i>E. coli</i> with LuxI secreted the 3OC6 HSL signal molecule, and the <i>E. coli</i> with LuxR received the signal and regulate the expression of downstream sfGFP, making it express. So we can detect green fluorescence in the latter <i>E. coli</i> under the irradiation of ultraviolet light. | + | We encapsulated <i>E. coli</i> with this composite part and <i>E. coli</i> with LuxR and sfGFP into alginate gel beads. The <i>E. coli</i> with LuxI secreted the 3OC6 HSL signal molecule, and the <i>E. coli</i> with LuxR received the signal and regulate the expression of downstream sfGFP, making it express. So we can detect green fluorescence in the latter <i>E. coli</i> under the irradiation of ultraviolet light and gain Figure 2. |
| − | [[File:LuxR regulations between E.coli.jpg| | + | [[File:LuxR regulations between E.coli.jpg|700px|thumb|left|'''Figure 2:''' sfGFP is expressed under the regulation of LuxR between <i>E. coli</i> in a single gel bead. Two parallel groups are given and the above two are control groups.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| − | + | We put the gel beads of the experimental group and the control group on a confocal microscope for imaging observation, and found that there was a significant difference in fluorescence intensity between the two groups (Figure 3-4). Thus, the Lux system was proved to be functional between <i>E. coli</i>.<br> | |
| − | + | [[File:SfGFP in gel beads Lux-.jpg|430px|thumb|left|'''Figure 3:''' 10X Confocal image of the control group's gel beads. ]] | |
| + | [[File:SfGFP in gel beads Lux+.jpg|430px|thumb|'''Figure 4:''' 10X Confocal image of the experimental group's gel beads. ]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| − | [[File:LuxI supernatant induces LuxR.jpg|500px|thumb|left|'''Figure | + | ==Quantitative analysis== |
| + | |||
| + | ===Experiment approach=== | ||
| + | 1. Centrifuge the overnight <i>E. coli</i> culture transformed with BBa_K4115040(express LuxI consititutively) at 12000 rpm for 1 min. Take out the supernatants for use.<br> | ||
| + | 2. Dilute the overnight <i>E. coli</i> culture that transformed with BBa_K4115039 to OD600~0.1.<br> | ||
| + | 3. Three groups are set: 1) Blank: diluted cultures in step2, autoinducer is not supplied to blank group 2) 10<sup>-7</sup> M 3OC6 HSL: 1:1000 add the 10<sup>-3</sup> M autoinducer into the diluted cultures, the final autoinducer concentration is 10<sup>-7</sup> M 3) Supernatant: centrifuge the diluted cultures at 3500 rpm for 10 min, resuspend the cells with the supernatant in step1. Culture all these three groups at 37℃ with shaking at 220rpm for 3h.<br> | ||
| + | 4. After three hours, add 200ul to each well of the black bottom 96-well plate for fluorescence intensity testing. Use an excitation wavelength of 485nm and an emission wavelength of 535nm.<br> | ||
| + | 5. Add 200ul to each well of the transparent 96-well plate for OD600 measurement. <br> | ||
| + | |||
| + | ===Results=== | ||
| + | |||
| + | We process the raw data by dividing the fluorescence intensity by the OD600 and get Figure 5. Thus, we can conclude the response intensity of LuxR to 3O6C HSL produced by BBa_K4115040. | ||
| + | |||
| + | [[File:LuxI supernatant induces LuxR.jpg|500px|thumb|left|'''Figure 5:''' LuxI supernatant induces LuxR to respond and express sfGFP.The test group is compared with 10<sup>-7</sup>M 3OC6 HSL added group and the blank group that only has the <i>E. coli</i> culture.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K4115040 SequenceAndFeatures</partinfo> | <partinfo>BBa_K4115040 SequenceAndFeatures</partinfo> | ||
| + | |||
== References == | == References == | ||
<small> | <small> | ||
Latest revision as of 15:03, 12 October 2022
J23101-B0034-LuxI-B0015
| J23101-B0034-LuxI-B0015 | |
|---|---|
| Function | Sythase |
| Use in | E. coli |
| RFC standard | RFC 10, RFC 10 compatible |
| Backbone | pUC57 |
| Submitted by | ShanghaiTech_China |
This composite part is used to produce 3OC6 HSL by LacI (BBa_C0161)[1] under a constitutive promoter J23101(BBa_J23101) and send it to the receiver protein LuxR (BBa_C0062)in the other E. coli.When the LuxR receives the signal, it will regulate and express the downstream report gene sfGFP [2], Figure 1. Thus we can detect whether LuxI is functional by observing the fluorescence intensity under UV light.
Proof of function
We encapsulated E. coli with this composite part and E. coli with LuxR and sfGFP into alginate gel beads. The E. coli with LuxI secreted the 3OC6 HSL signal molecule, and the E. coli with LuxR received the signal and regulate the expression of downstream sfGFP, making it express. So we can detect green fluorescence in the latter E. coli under the irradiation of ultraviolet light and gain Figure 2.
We put the gel beads of the experimental group and the control group on a confocal microscope for imaging observation, and found that there was a significant difference in fluorescence intensity between the two groups (Figure 3-4). Thus, the Lux system was proved to be functional between E. coli.
Quantitative analysis
Experiment approach
1. Centrifuge the overnight E. coli culture transformed with BBa_K4115040(express LuxI consititutively) at 12000 rpm for 1 min. Take out the supernatants for use.
2. Dilute the overnight E. coli culture that transformed with BBa_K4115039 to OD600~0.1.
3. Three groups are set: 1) Blank: diluted cultures in step2, autoinducer is not supplied to blank group 2) 10-7 M 3OC6 HSL: 1:1000 add the 10-3 M autoinducer into the diluted cultures, the final autoinducer concentration is 10-7 M 3) Supernatant: centrifuge the diluted cultures at 3500 rpm for 10 min, resuspend the cells with the supernatant in step1. Culture all these three groups at 37℃ with shaking at 220rpm for 3h.
4. After three hours, add 200ul to each well of the black bottom 96-well plate for fluorescence intensity testing. Use an excitation wavelength of 485nm and an emission wavelength of 535nm.
5. Add 200ul to each well of the transparent 96-well plate for OD600 measurement.
Results
We process the raw data by dividing the fluorescence intensity by the OD600 and get Figure 5. Thus, we can conclude the response intensity of LuxR to 3O6C HSL produced by BBa_K4115040.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[1] Daer R, Barrett CM, Melendez EL, Wu J, Tekel SJ, Xu J, Dennison B, Muller R, Haynes KA. Characterization of diverse homoserine lactone synthases in Escherichia coli. PLoS One. 2018 Aug 23;13(8):e0202294. doi: 10.1371/journal.pone.0202294. PMID: 30138364; PMCID: PMC6107141.
[2] Kylilis, N., Tuza, Z.A., Stan, GB. et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia. Nat Commun 9, 2677 (2018). https://doi.org/10.1038/s41467-018-05046-2