Difference between revisions of "Part:BBa K3757007"
Erik zimmer (Talk | contribs) (edited part number) |
Erik zimmer (Talk | contribs) |
||
| Line 5: | Line 5: | ||
Gene encoding the cyclotide precursor Oak1 (BBa_K3757001), C-terminally tagged with a c-myc tag (BBa_K3757008). Regulated by the eukaryotic constitutive CaMV 35S promoter (BBa_K788000) and the 35S terminator polyadenylation signal (BBa_K1159307). In one Golden Gate cloning step, a sequence encoding a peptide can be cloned into the Oak1 precursor. This gene can then be co-expressed with CtAEP1 in Nicotiana benthamiana leading to expression of the peptide in cyclized form. | Gene encoding the cyclotide precursor Oak1 (BBa_K3757001), C-terminally tagged with a c-myc tag (BBa_K3757008). Regulated by the eukaryotic constitutive CaMV 35S promoter (BBa_K788000) and the 35S terminator polyadenylation signal (BBa_K1159307). In one Golden Gate cloning step, a sequence encoding a peptide can be cloned into the Oak1 precursor. This gene can then be co-expressed with CtAEP1 in Nicotiana benthamiana leading to expression of the peptide in cyclized form. | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3757007 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3757007 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | </p> | ||
| + | </html> | ||
| + | __TOC__ | ||
| + | |||
| + | |||
| + | ===Usage and Biology=== | ||
| + | |||
| + | This part can be used for the constitutive expression of a grafted cyclotide construct embedded in the Oak1 precursor in plants, C-terminally tagged with a c-myc-tag. After expression, the included tag may be used for affinity purification or detection of the precursor with an anti-c-myc-tag antibody in an immunoassay. It inherits the following basic parts: | ||
| + | * CaMV 35S promoter (<partinfo>BBa_K788000</partinfo>) | ||
| + | * Oak1 precursor (<partinfo>BBa_K3757001</partinfo>) | ||
| + | * c-myc-tag (<partinfo>BBa_K3757008</partinfo>) | ||
| + | * 35S terminator (<partinfo>BBa_K1159307</partinfo>) | ||
| + | |||
| + | |||
| + | ===Cloning=== | ||
| + | |||
| + | The correct cloning of the cyclotide with the respective AMP into the Oak1 precursor in our empty 3in1 vector (<partinfo>BBa_K3757011</partinfo>) could also be verified through blue-white screening of the grown colonies on LB-agar plates supplemented with X-Gal and Isopropyl-β-D-thiogalactopyranoside (IPTG) (figure 2). Hence, we could recognize successfully cloned colonies by their color because bacteria carrying the 3in1 plasmid with an AMP construct in their Oak1 precursor appeared white and not blue due to them not expressing β-galactosidase. | ||
| + | |||
| + | [[Image:T--Tuebingen--results_cloning_Fig2.jpeg|700px|thumb|center|'''Figure 2: Blue-white screening.''' Blue-white screening enabled us to distinguish between empty level III (blue colonies) and level IV vectors (white colonies). The fact, that there are far more white than blue colonies showed us, that our Golden Gate assembly protocol is very efficient.]] | ||
| + | |||
| + | |||
| + | ===Expression in <i>Nicotiana benthamiana</i>=== | ||
| + | |||
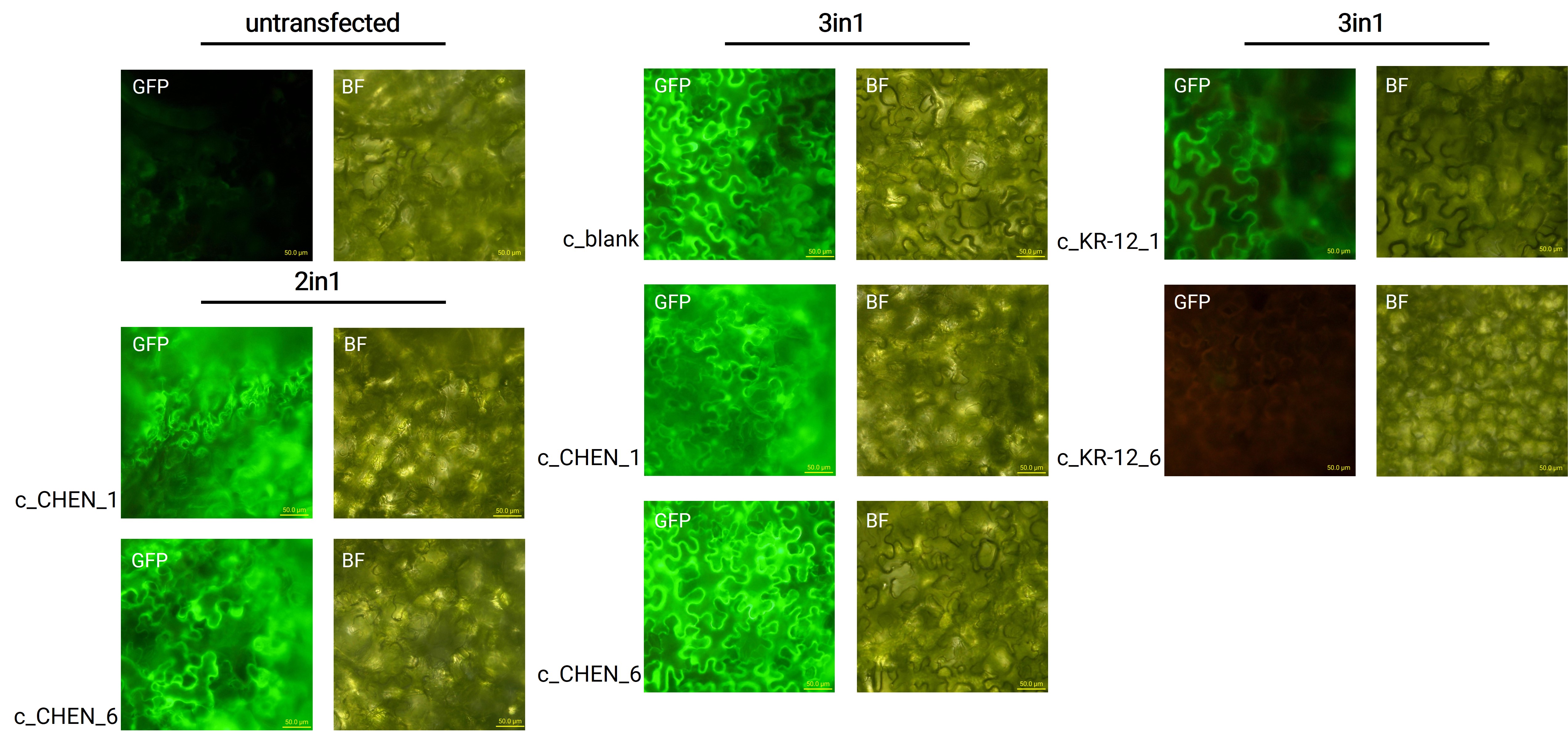
| + | Oak1 precursor containing an AMP encoding sequence was co-expressed with GFP (<partinfo>BBa_K3669012</partinfo>) as a reporter gene. Four days after agroinfiltration of <i>N. benthamiana</i> leaves, leaf pieces were analysed with fluorescence microscopy (figure 3) to confirm GFP expression. In comparison to the negative control, leaves transfected with our final vectors showed clear green fluorescence for all except the c_KR-12_6 construct. As a negative control, non-infiltrated leaves or leaves infiltrated with p19 only were checked for green fluorescence. As shown in figure 3, this negative control showed no significant level of green fluorescence, confirming that the green fluorescence we detected for other vectors can only result from successful expression of one of our final vectors. | ||
| + | |||
| + | [[Image:T--Tuebingen--results_expression_fluoro_figure2.jpeg|700px|thumb|center|'''Figure 3: Microscopy images from untransfected N. benthamiana leaves and leaves infiltrated with the respective 2in1 and 3in1 vectors.''' GFP – fluorescence channel, BF – brightfield microscopy, scale bar – 50.0 µm.]] | ||
| + | |||
| + | |||
| + | We can draw the conclusion that the transfection was successful for both the 2in1 and the 3in1 vectors. However, not the whole infiltrated leaves were fluorescent but defined loci only, which indicates a low expression rate. | ||
| + | |||
| + | |||
| + | ===Extraction from <i>Nicotiana benthamiana</i>=== | ||
| + | |||
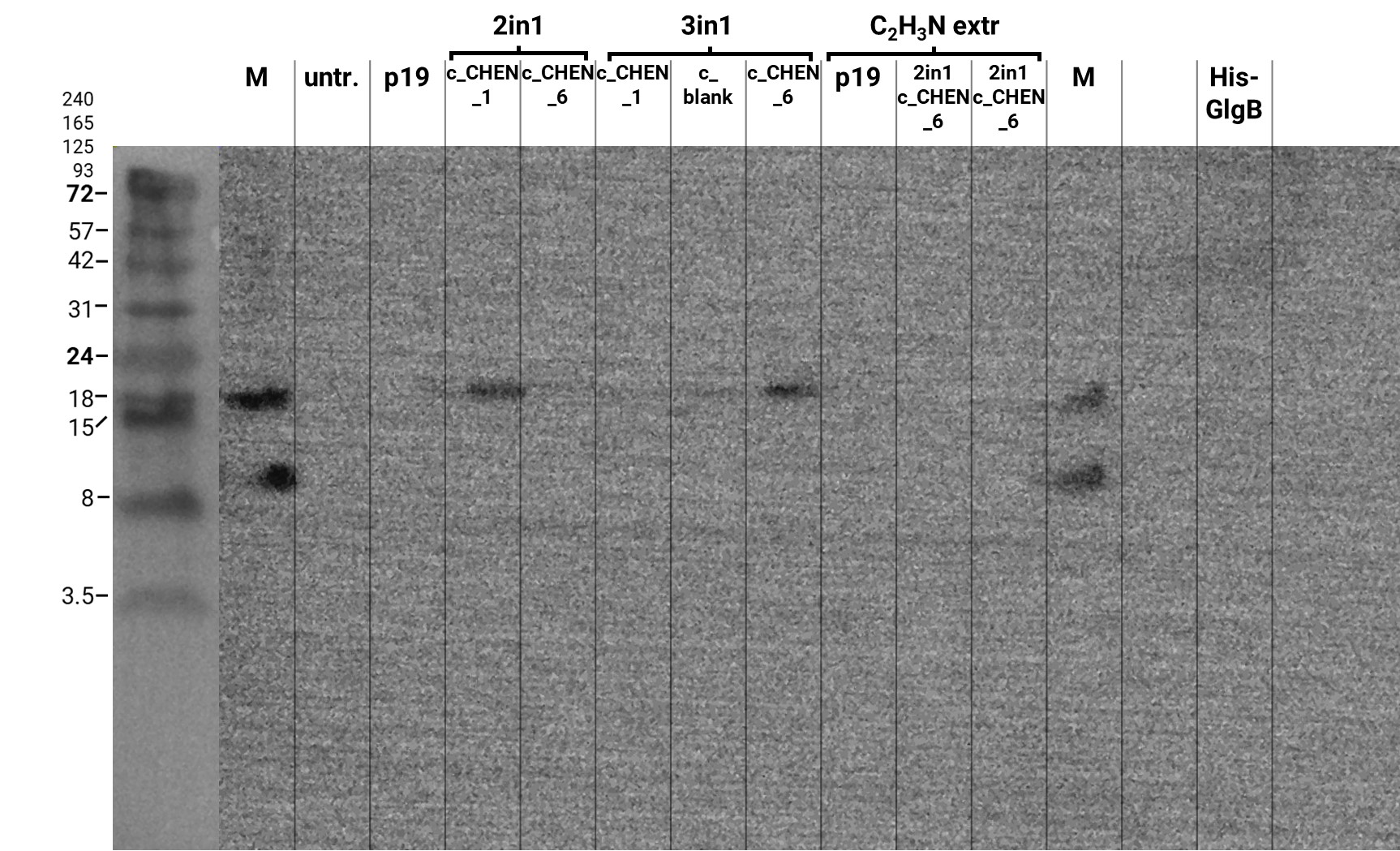
| + | We performed a western blot of the plant crude extract samples, with an anti-His-tag primary antibody to be able to specifically detect our His-tagged cyclotides. An immunoblot signal at a size of about 18 kDa could be detected (figure 4), which raised the suspicion that cyclization of our cyclotides by the AEP did not work, as the Oak1 precursor of our constructs is about the same size as the detected signal. Another possible explanation is that the band shows indeed our cyclotide, which does not run at the expected molecular range in the SDS-PAGE. This could be due to its highly positive charge, which might not be completely masked by the negatively charged SDS or due to its cyclic structure. Unfortunately, no signal was detected for our positive control, a His-tagged protein of about 84 kDa. This could be explained by the huge size difference between this control and our peptide. Thus, it could have happened that the blotting of the control on the membrane did not work. | ||
| + | |||
| + | [[Image:T--Tuebingen--results_western_antiHis_2021_09_29_5min.jpeg|600px|thumb|center|'''Figure 4: Results of the western blot for specific detection of His-tagged proteins.''' The separated protein bands on the SDS-PAGE gel of the samples extracted via Lämmli or acetonitrile-based buffer were blotted onto a PVDF membrane for further analysis. The membrane was blocked in 5% milk/TBS-T and afterward O.N. incubated with an anti-His-tag primary antibody. After incubation with the corresponding secondary antibody, the signal was detected after five minutes of exposure time. M-marker, untr.-untransfected, p19-control vector, 2in1 and 3in1 constructs labeled according to our official nomenclature, His-GlgB-positive control (~ 84 kDa).]] | ||
Revision as of 01:04, 22 October 2021
35S Oak1
Gene encoding the cyclotide precursor Oak1 (BBa_K3757001), C-terminally tagged with a c-myc tag (BBa_K3757008). Regulated by the eukaryotic constitutive CaMV 35S promoter (BBa_K788000) and the 35S terminator polyadenylation signal (BBa_K1159307). In one Golden Gate cloning step, a sequence encoding a peptide can be cloned into the Oak1 precursor. This gene can then be co-expressed with CtAEP1 in Nicotiana benthamiana leading to expression of the peptide in cyclized form.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1468
Illegal BamHI site found at 1858 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 66
Contents
Usage and Biology
This part can be used for the constitutive expression of a grafted cyclotide construct embedded in the Oak1 precursor in plants, C-terminally tagged with a c-myc-tag. After expression, the included tag may be used for affinity purification or detection of the precursor with an anti-c-myc-tag antibody in an immunoassay. It inherits the following basic parts:
- CaMV 35S promoter (BBa_K788000)
- Oak1 precursor (BBa_K3757001)
- c-myc-tag (BBa_K3757008)
- 35S terminator (BBa_K1159307)
Cloning
The correct cloning of the cyclotide with the respective AMP into the Oak1 precursor in our empty 3in1 vector (BBa_K3757011) could also be verified through blue-white screening of the grown colonies on LB-agar plates supplemented with X-Gal and Isopropyl-β-D-thiogalactopyranoside (IPTG) (figure 2). Hence, we could recognize successfully cloned colonies by their color because bacteria carrying the 3in1 plasmid with an AMP construct in their Oak1 precursor appeared white and not blue due to them not expressing β-galactosidase.
Expression in Nicotiana benthamiana
Oak1 precursor containing an AMP encoding sequence was co-expressed with GFP (BBa_K3669012) as a reporter gene. Four days after agroinfiltration of N. benthamiana leaves, leaf pieces were analysed with fluorescence microscopy (figure 3) to confirm GFP expression. In comparison to the negative control, leaves transfected with our final vectors showed clear green fluorescence for all except the c_KR-12_6 construct. As a negative control, non-infiltrated leaves or leaves infiltrated with p19 only were checked for green fluorescence. As shown in figure 3, this negative control showed no significant level of green fluorescence, confirming that the green fluorescence we detected for other vectors can only result from successful expression of one of our final vectors.
We can draw the conclusion that the transfection was successful for both the 2in1 and the 3in1 vectors. However, not the whole infiltrated leaves were fluorescent but defined loci only, which indicates a low expression rate.
Extraction from Nicotiana benthamiana
We performed a western blot of the plant crude extract samples, with an anti-His-tag primary antibody to be able to specifically detect our His-tagged cyclotides. An immunoblot signal at a size of about 18 kDa could be detected (figure 4), which raised the suspicion that cyclization of our cyclotides by the AEP did not work, as the Oak1 precursor of our constructs is about the same size as the detected signal. Another possible explanation is that the band shows indeed our cyclotide, which does not run at the expected molecular range in the SDS-PAGE. This could be due to its highly positive charge, which might not be completely masked by the negatively charged SDS or due to its cyclic structure. Unfortunately, no signal was detected for our positive control, a His-tagged protein of about 84 kDa. This could be explained by the huge size difference between this control and our peptide. Thus, it could have happened that the blotting of the control on the membrane did not work.