Difference between revisions of "Part:BBa K592015"
| (9 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K592015 short</partinfo> | <partinfo>BBa_K592015 short</partinfo> | ||
| − | This color expressing cassette is a potential alternative to BBa_J04450, the current RFP cassette in all of the BioBrick backbones. Bacterial colonies exhibit strong blue color after 18 hours. Due to the much darker blue/purple color tone, even partially blue colonies are readily recognized. | + | This color expressing cassette is a potential alternative/complementary to <partinfo>BBa_J04450</partinfo>, the current RFP cassette in all of the BioBrick backbones. Bacterial colonies exhibit strong blue color after 18 hours. Due to the much darker blue/purple color tone, even partially blue colonies are readily recognized. |
| − | We found this design very useful when assembling components that contain RFP, mCherry, etc. The assembled parts are not recognizable from the background insert if all colonies becomes red after transformation. Luckily, very few genes produce substances that make the cells turn blue/purple, which makes our construct useful. | + | We found this design very useful when assembling components that contain RFP, mCherry, etc. The assembled parts are not recognizable from the background insert if all colonies becomes red after transformation. Luckily, very few genes produce substances that make the cells turn blue/purple, which makes our construct useful. Due to the strong promoter, over-expressing amilCP will be costly to the cells. |
| − | The promoter is constitutive, the RBS is strong, the double terminator is not | + | The promoter is constitutive, the RBS is strong, the double terminator is not <partinfo>BBa_B0015</partinfo> but <partinfo>BBa_B0014</partinfo>. |
[[Image:AmilCP_amilGFP_RFP.jpg|300px]] | [[Image:AmilCP_amilGFP_RFP.jpg|300px]] | ||
| Line 12: | Line 12: | ||
[[Image:Pellets BYR.jpg|300px]] | [[Image:Pellets BYR.jpg|300px]] | ||
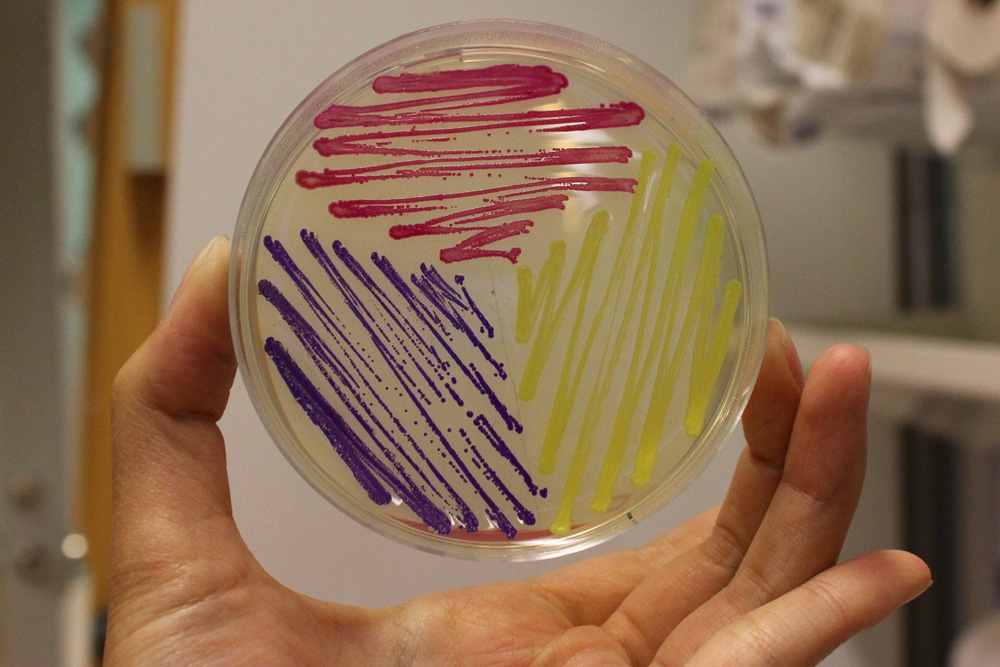
| − | The images above show ''E coli'' constitutively expressing amilCP <partinfo>BBa_K592009</partinfo> (blue), amilGFP BBa_K592010 (yellow), and RFP <partinfo>BBa_E1010</partinfo> (red). | + | The images above show ''E coli'' constitutively expressing amilCP <partinfo>BBa_K592009</partinfo> (blue), amilGFP <partinfo>BBa_K592010</partinfo> (yellow), and RFP <partinfo>BBa_E1010</partinfo> (red). |
| + | <br/> | ||
| + | <br/> | ||
| + | =Improvement by ICJFLS2021= | ||
| + | ==Overview:== | ||
| + | <br/> | ||
| + | BBa_K592015 is a amilCP blue coding device, which consists of J23101, B0034, K592009, and B0014. It was constructed by iGEM11_Uppsala-Sweden in Sep. 2011. This device can express blue/purple color protein, which enriches the reporter genes for the research of life science. K592009 is a blue chromo protein gene from the Acropora millepora. AmilCP, naturally exhibits strong color when expressed. The protein has an absorbance maximum at 588 nm giving it a blue/purple color visible to the naked eye, thereby requiring no instruments to observe. The strong color is readily observed in both LB or agar culture, in less than 24 hours of incubation. It is a very useful part. | ||
| + | <br/> | ||
| + | However, the promoter (J23101) of this part is a constitutive strong promoter, and the RBS (B0034) is strong too, making amilCP over-expressed in the cells. This is a burden and costly to the transformed cells. | ||
| + | <br/> | ||
| + | Plac is one of the most common promoters, and it is negatively regulated by LacI repressor. Serving as inducer, IPTG can bind with LacI repressor, which enables the transcription of downstream genes. Although the E. coli could express some LacI, it is not enough for repression the transformed promoter Plac in E.coli. For example, BBa_J04450 is RFP generator driven by Plac promoter. This RFP generator has some leakage expression, as the designer stated that some RFP express even without IPTG inducer presence. | ||
| + | <br/> | ||
| + | Based on the above description, we constructed a new amilCP device (BBa_K3867009) containing LacI gene, which is driven by Plac promoter. Both LacI and amilCP are connected to the downstream of Plac, which is regulated by both LacI repressor and IPTG inducer. This new amilCP device could be self-regulated because LacI protein can inhibit its self-expression. When amilCP needs expression, IPTG is required to be added into the culture. This induction expression could be considered as an economic expression way. Using the absorbance at 588nm (the maximum wave length of amilCP), we detected the response of this device to different concentration of IPTG, indicating that it could be inhibited by LacI, and induced well by IPTG. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | ==Results:== | ||
| + | <br/> | ||
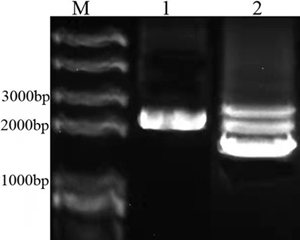
| + | In order to construct BBa_K3867009, overlapping PCR method was used to amplify the genes of Plac, LacI and BBa_K592015. Then the PCR product was inserted into plasmid pSB1C3 to create the new part BBa_K3867009. The identification result is showed in Fig.1. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | [[File:K3867009-6.jpg|center]] | ||
| + | <br/> | ||
| + | Fig.1. The identification result of BBa_K3867009 construction. M: Marker; 1: PCR result of amilCP device from BBa_K3867009; 2: Plasmid of BBa_K3867009. | ||
| + | <br/> | ||
| + | <br/> | ||
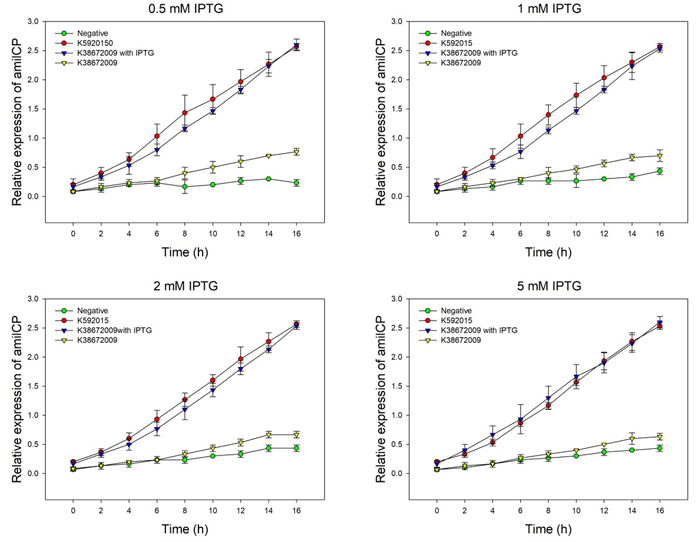
| + | We detected the effect of IPTG induction on the two amilCP devices, using different concentration of IPTG. We set 4 groups: the original amilCP device (BBa_K592015), the new amilCP device (BBa_K3867009) with or without IPTG, and one negative control without amilCP expression. When all groups’ OD600 approximately reached to 0.6, a certain concentration of IPTG was added to the culture medium, incubated cells at 37℃ for 16h. Measure the absorbance values at 588 nm and 600 nm for each group every 2h, using an automatic microplate reader. The results are showed as follows (Fig.2-Fig.3). | ||
| + | <br/> | ||
| + | <br/> | ||
| + | [[File:K3867009-8.jpg|center]] | ||
| + | <br/> | ||
| + | Fig.2. The amilCP expression of BBa_K592015 and BBa_K3867009 with and without IPTG induction. The relative expression of amilCP is standardized with the ratio of OD588 value to the OD600 value for each group every 2h. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | The figure showed that both BBa_K592015 and BBa_K3867009 expressed amilCP, and different concentration of IPTG had the same inducing trend for K3867009. The results indicated that BBa_K592015 expressed a mountain of amilCP without IPTG. However, BBa_K3867009 expressed a high level of amilCP only with IPTG presence. The leakage expression of amilCP is very low, and it is sensitive to the IPTG induction. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | [[File:K3867009-5.jpg|center]] | ||
| + | <br/> | ||
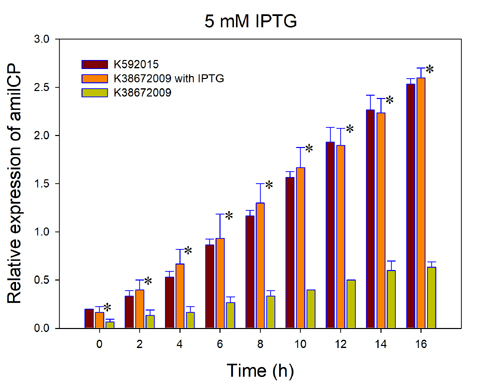
| + | Fig.3. The comparison of amilCP expressed by BBa_K592015 and BBa_K3867009 with and without IPTG induction. The relative expression of amilCP is standardized with the ration of OD588 value to OD600 value for each group every 2h. “*”, significant difference compared to k38672009 without IPTG induction. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | This figure indicated that the original amilCP device (BBa_K592015) can express amilCP with a high constant level. However, the new part amilCP device BBa_K3867009 express a high level of amilCP only with the IPTG induction, which is considered as an economic way. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
| + | |||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K592015 SequenceAndFeatures</partinfo> | <partinfo>BBa_K592015 SequenceAndFeatures</partinfo> | ||
Latest revision as of 21:35, 21 October 2021
amilCP Blue Coding Device
This color expressing cassette is a potential alternative/complementary to BBa_J04450, the current RFP cassette in all of the BioBrick backbones. Bacterial colonies exhibit strong blue color after 18 hours. Due to the much darker blue/purple color tone, even partially blue colonies are readily recognized.
We found this design very useful when assembling components that contain RFP, mCherry, etc. The assembled parts are not recognizable from the background insert if all colonies becomes red after transformation. Luckily, very few genes produce substances that make the cells turn blue/purple, which makes our construct useful. Due to the strong promoter, over-expressing amilCP will be costly to the cells.
The promoter is constitutive, the RBS is strong, the double terminator is not BBa_B0015 but BBa_B0014.
The images above show E coli constitutively expressing amilCP BBa_K592009 (blue), amilGFP BBa_K592010 (yellow), and RFP BBa_E1010 (red).
Improvement by ICJFLS2021
Overview:
BBa_K592015 is a amilCP blue coding device, which consists of J23101, B0034, K592009, and B0014. It was constructed by iGEM11_Uppsala-Sweden in Sep. 2011. This device can express blue/purple color protein, which enriches the reporter genes for the research of life science. K592009 is a blue chromo protein gene from the Acropora millepora. AmilCP, naturally exhibits strong color when expressed. The protein has an absorbance maximum at 588 nm giving it a blue/purple color visible to the naked eye, thereby requiring no instruments to observe. The strong color is readily observed in both LB or agar culture, in less than 24 hours of incubation. It is a very useful part.
However, the promoter (J23101) of this part is a constitutive strong promoter, and the RBS (B0034) is strong too, making amilCP over-expressed in the cells. This is a burden and costly to the transformed cells.
Plac is one of the most common promoters, and it is negatively regulated by LacI repressor. Serving as inducer, IPTG can bind with LacI repressor, which enables the transcription of downstream genes. Although the E. coli could express some LacI, it is not enough for repression the transformed promoter Plac in E.coli. For example, BBa_J04450 is RFP generator driven by Plac promoter. This RFP generator has some leakage expression, as the designer stated that some RFP express even without IPTG inducer presence.
Based on the above description, we constructed a new amilCP device (BBa_K3867009) containing LacI gene, which is driven by Plac promoter. Both LacI and amilCP are connected to the downstream of Plac, which is regulated by both LacI repressor and IPTG inducer. This new amilCP device could be self-regulated because LacI protein can inhibit its self-expression. When amilCP needs expression, IPTG is required to be added into the culture. This induction expression could be considered as an economic expression way. Using the absorbance at 588nm (the maximum wave length of amilCP), we detected the response of this device to different concentration of IPTG, indicating that it could be inhibited by LacI, and induced well by IPTG.
Results:
In order to construct BBa_K3867009, overlapping PCR method was used to amplify the genes of Plac, LacI and BBa_K592015. Then the PCR product was inserted into plasmid pSB1C3 to create the new part BBa_K3867009. The identification result is showed in Fig.1.
Fig.1. The identification result of BBa_K3867009 construction. M: Marker; 1: PCR result of amilCP device from BBa_K3867009; 2: Plasmid of BBa_K3867009.
We detected the effect of IPTG induction on the two amilCP devices, using different concentration of IPTG. We set 4 groups: the original amilCP device (BBa_K592015), the new amilCP device (BBa_K3867009) with or without IPTG, and one negative control without amilCP expression. When all groups’ OD600 approximately reached to 0.6, a certain concentration of IPTG was added to the culture medium, incubated cells at 37℃ for 16h. Measure the absorbance values at 588 nm and 600 nm for each group every 2h, using an automatic microplate reader. The results are showed as follows (Fig.2-Fig.3).
Fig.2. The amilCP expression of BBa_K592015 and BBa_K3867009 with and without IPTG induction. The relative expression of amilCP is standardized with the ratio of OD588 value to the OD600 value for each group every 2h.
The figure showed that both BBa_K592015 and BBa_K3867009 expressed amilCP, and different concentration of IPTG had the same inducing trend for K3867009. The results indicated that BBa_K592015 expressed a mountain of amilCP without IPTG. However, BBa_K3867009 expressed a high level of amilCP only with IPTG presence. The leakage expression of amilCP is very low, and it is sensitive to the IPTG induction.
Fig.3. The comparison of amilCP expressed by BBa_K592015 and BBa_K3867009 with and without IPTG induction. The relative expression of amilCP is standardized with the ration of OD588 value to OD600 value for each group every 2h. “*”, significant difference compared to k38672009 without IPTG induction.
This figure indicated that the original amilCP device (BBa_K592015) can express amilCP with a high constant level. However, the new part amilCP device BBa_K3867009 express a high level of amilCP only with the IPTG induction, which is considered as an economic way.