Difference between revisions of "Part:BBa K3740033"
(→2021 SZPT-China) |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3740033 short</partinfo> | <partinfo>BBa_K3740033 short</partinfo> | ||
| + | ===Description=== | ||
| + | This composite part is a generator consisting of the pDawn promoter and LKD16. | ||
| + | ===Sequence and Features=== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<partinfo>BBa_K3740033 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3740033 SequenceAndFeatures</partinfo> | ||
| Line 17: | Line 13: | ||
<partinfo>BBa_K3740033 parameters</partinfo> | <partinfo>BBa_K3740033 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | =2021 SZPT-China= | ||
| + | <h3>Biology</h3> | ||
| + | <p>This composite part is a generator consisting of the pDawn (<partinfo>BBa_K1075044</partinfo>) promoter and LKD16 (<partinfo>BBa_K3740032</partinfo>).</p> | ||
| + | <h3>Usage</h3> | ||
| + | <p>The target gene of pDawn blue light response system (<partinfo>BBa_K1075044</partinfo>) and Lysis Cassette LKD16 (<partinfo>BBa_K3740032</partinfo>) were insert into the pSEVA331 expression vector. Then, random primer guided mutagenesis method to modulate the strength of the RBS (<partinfo>BBa_B0034</partinfo>) located upstream of LKD16. After introducing into <i>E. coli</i> DH5α, <i>E. coli</i> isolates that normally grow in the dark and cannot under the light of 470nm blue light were screened. Finally, the pDawn-RBSNNN-LKD16-rrnB T1 plasmid was extracted, and further introduced into <i>G. hansenii</i> ATCC 53582, in which the responsiveness of pDawn to blue light was also verified.</p> | ||
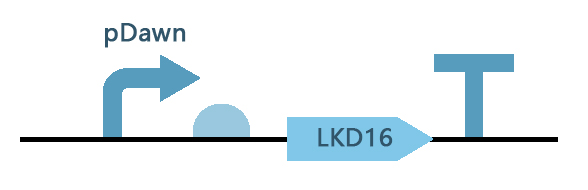
| + | [[File:szpt.90.jpg|600px|thumb|center|Figure 1. Gene circuit of LKD16(<partinfo>BBa_K3740032</partinfo>).]] | ||
| + | <h3>Characterization</h3> | ||
| + | <h4>1.Batch screening of pDawn-RBSNNN-LKD16-rrnB T1-pSEVA331 in response to blue light lysis in <i>E. coli</i>.</h4> | ||
| + | <p>As shown in Figure 2, the 4<sup>th</sup>, 9<sup>th</sup>, 10<sup>th</sup> and 12<sup>th</sup> of the plate can grow normally in the dark, but not grow under blue light, <b>indicating that LKD16 can induce lysis in <i>E. coli</i> in response to blue light.</b></p> | ||
| + | [[File:szpt40.png|600px|thumb|center|Figure 2. The growth status of pDawn-RBSNNN-rrnB T1-LKD16-pSEVA331-DH5α under blue light (a) and dark conditions (b).]] | ||
| + | <h4>2. pDawn-RBSNNN-LKD16-rrnB T1-pSEVA331-8# in response to blue photolysis in <i>G. hansenii</i> ATCC 53582.</h4> | ||
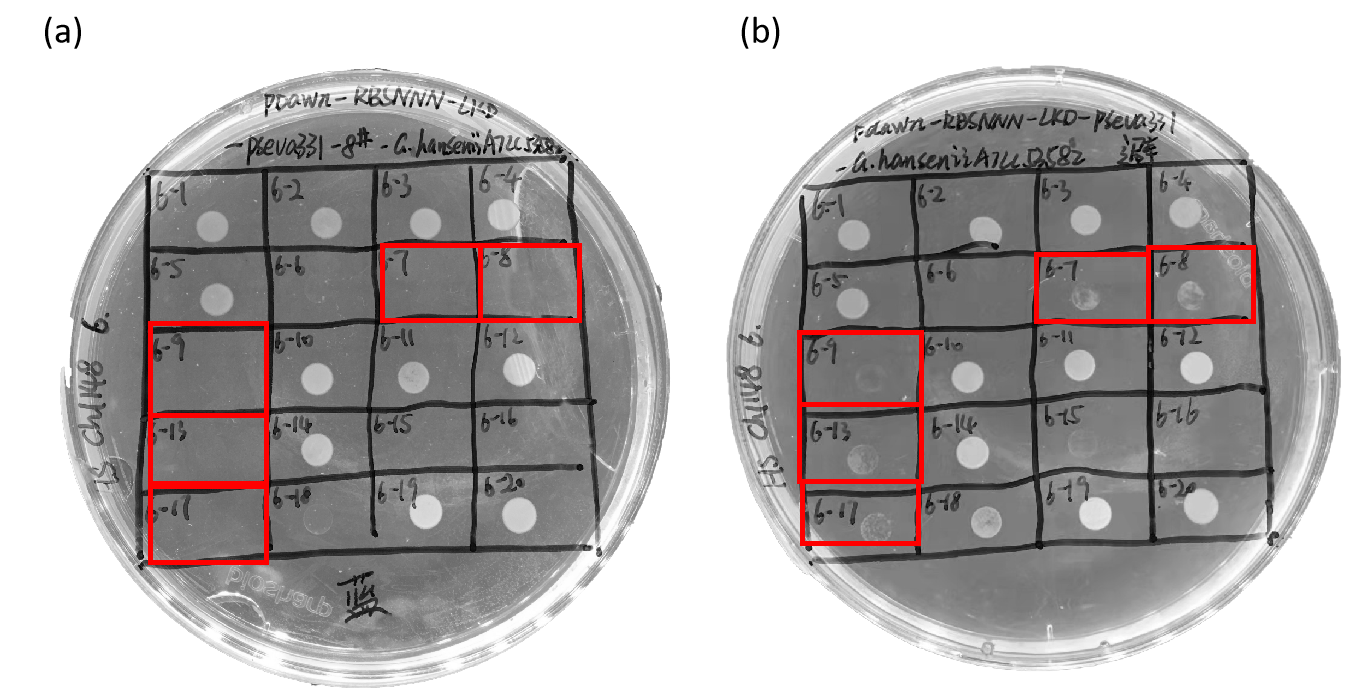
| + | <p>As shown in Figure 3, although 7<sup>th</sup>, 8<sup>th</sup>, 9<sup>th</sup>, 13<sup>th</sup> and 17<sup>th</sup> on the plate can grow under the dark, not grow under blue light, the growth of the bacteria under dark conditions is abnormal. <b>Therefore, although the LKD16 cleavage protein has a cleavage function, its function is not very stable.</b></p> | ||
| + | [[File:szpt41.png|600px|thumb|center|Figure 3. pDawn-RBSNNN-LKD16-rrnB T1-pSEVA331-<i>G.hansenii</i> ATCC 53582-8# cultured under blue light (a) and dark conditions (b).]] | ||
| + | <h3>References</h3> | ||
| + | <p>[1] Ceyssens P J, Lavigne R, Mattheus W, et al. Genomic Analysis of Pseudomonas aeruginosa Phages LKD16 and LKA1: Establishment of the KMV Subgroup within the T7 Supergroup[J]. Journal of Bacteriology, 2006.</p> | ||
Latest revision as of 18:02, 21 October 2021
pDawn-B0034-LKD16-rrnB T1
Description
This composite part is a generator consisting of the pDawn promoter and LKD16.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2171
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 63
Illegal NgoMIV site found at 195
Illegal NgoMIV site found at 289
Illegal NgoMIV site found at 582
Illegal NgoMIV site found at 1076
Illegal NgoMIV site found at 1094
Illegal NgoMIV site found at 1184
Illegal AgeI site found at 414
Illegal AgeI site found at 1542 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1643
Illegal BsaI.rc site found at 525
2021 SZPT-China
Biology
This composite part is a generator consisting of the pDawn (BBa_K1075044) promoter and LKD16 (BBa_K3740032).
Usage
The target gene of pDawn blue light response system (BBa_K1075044) and Lysis Cassette LKD16 (BBa_K3740032) were insert into the pSEVA331 expression vector. Then, random primer guided mutagenesis method to modulate the strength of the RBS (BBa_B0034) located upstream of LKD16. After introducing into E. coli DH5α, E. coli isolates that normally grow in the dark and cannot under the light of 470nm blue light were screened. Finally, the pDawn-RBSNNN-LKD16-rrnB T1 plasmid was extracted, and further introduced into G. hansenii ATCC 53582, in which the responsiveness of pDawn to blue light was also verified.
Characterization
1.Batch screening of pDawn-RBSNNN-LKD16-rrnB T1-pSEVA331 in response to blue light lysis in E. coli.
As shown in Figure 2, the 4th, 9th, 10th and 12th of the plate can grow normally in the dark, but not grow under blue light, indicating that LKD16 can induce lysis in E. coli in response to blue light.
2. pDawn-RBSNNN-LKD16-rrnB T1-pSEVA331-8# in response to blue photolysis in G. hansenii ATCC 53582.
As shown in Figure 3, although 7th, 8th, 9th, 13th and 17th on the plate can grow under the dark, not grow under blue light, the growth of the bacteria under dark conditions is abnormal. Therefore, although the LKD16 cleavage protein has a cleavage function, its function is not very stable.
References
[1] Ceyssens P J, Lavigne R, Mattheus W, et al. Genomic Analysis of Pseudomonas aeruginosa Phages LKD16 and LKA1: Establishment of the KMV Subgroup within the T7 Supergroup[J]. Journal of Bacteriology, 2006.