Difference between revisions of "Part:BBa K3886003"
| Line 7: | Line 7: | ||
===Usage and Biology/Characterisation=== | ===Usage and Biology/Characterisation=== | ||
| − | + | ||
| − | + | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="right" | |
| − | + | ! colspan="2" style="background:#FFBF00;"|CMV:Ha-NLS-dCas9-Linker-VP16-NLS:BGH | |
| − | + | |- | |
| + | |'''Function''' | ||
| + | |gene activation | ||
| + | |- | ||
| + | |'''Use in''' | ||
| + | |Prokaryotic cells | ||
| + | |- | ||
| + | |'''RFC standard''' | ||
| + | |[https://parts.igem.org/Help:Assembly_standard_10 RFC 10], RFC 10 compatible | ||
| + | |- | ||
| + | |'''Backbone''' | ||
| + | |pSB1C3<br> | ||
| + | |- | ||
| + | |'''Submitted by''' | ||
| + | |[http://2016.igem.org/Team:NEU-China] | ||
| + | |} | ||
| + | |||
| + | |||
| + | The Freiburg iGEM team 2013 designed a fusion protein consisting of dCas9 and VP16 [1-6] for sequence-specific transactivation of a desired target locus [http://2013.igem.org/Team:Freiburg/Project/effector#activation (more information)]. Therefore, we used our double mutated dCas9 [https://parts.igem.org/Part:BBa_K1150000 (BBa_K1150000)] impaired in its cleavage activity and fused it to the 5’ end of the sequence coding for the transactivation domain of VP16 [https://parts.igem.org/Part:BBa_K1150001 (BBa_K1150001)]. To ensure nuclear localization of the construct a nuclear localization signal (NLS) was fused to both ends of dCas9-VP16. For detection of expression the fusion protein was tagged with a HA-epitope coding sequence [https://parts.igem.org/Part:BBa_K1150016 (BBaa_K1150016)] and its expression was set under control of the CMV promoter [https://parts.igem.org/Part:BBa_K747096 (BBa_K747096)] and BGH terminator [https://parts.igem.org/Part:BBa_K1150012 (BBa_K1150012)]. Figure 1 illustrates the detailed design of the whole device. | ||
| + | |||
| + | [[File:Overview_of_the_construct_CMV-Cas-VP16_Freigem_2013.png|700px|]] | ||
| + | |||
| + | '''Figure 1: Construct design.''' dCas9 was fused via a 3 amino acid linker to VP16. The resulting fusion construct was flanked by NLS sequences and tagged by a HA epitope. The CMV promoter and BGH terminator were chosen to control gene expression. | ||
| + | |||
| + | By co-transfecting our RNA plasmid [https://parts.igem.org/Part:BBa_K1150034 (BBa_K1150034)] which includes the tracrRNA and the separately integrated, desired crRNA, the dCas9 specifically binds to the targeted DNA sequence. With the help of the transactivation domain of VP16, transcription factors are recruited and the pre-initiation complex can be built. By placing this construct upstream of a promotor region any gene of interest can be activated. | ||
| + | |||
| + | |||
| + | [[File:Picture1VP16-Cas_Freigem2013.png|700px|]] | ||
| + | |||
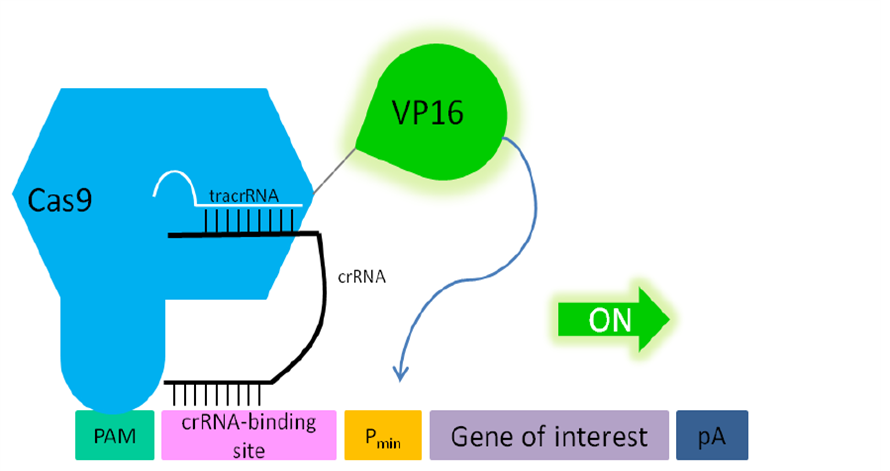
| + | '''Figure 2: Principle of transactivation of mammalian gene expression by the fusion protein dCas9-VP19 (BBa_K1150020).''' The double mutated dCas9 (D10A; H840A) fused to the herpes simplex virus (HSV) derived VP16 activation domain can serve as a crRNA-guided DNA-binding and transactivating protein. If a PAM sequence is present at the 3’ end of the crRNA binding site almost any DNA sequence can be targeted. Abbr.: Pmin: minimal promoter, containing minimal requirements for binding of transcription factors; Cas9: CRISPR associated protein 9; crRNA: CRISPR RNA; tracrRNA: trans-activating RNA; VP16: Virus protein 16, herpes simplex virus (HSV)-derived transcriptional activator protein; PAM: protospacer adjacent motif. | ||
Revision as of 20:54, 14 October 2021
Caffeine Sensor
A Caffeine Sensor
Usage and Biology/Characterisation
| CMV:Ha-NLS-dCas9-Linker-VP16-NLS:BGH | |
|---|---|
| Function | gene activation |
| Use in | Prokaryotic cells |
| RFC standard | RFC 10, RFC 10 compatible |
| Backbone | pSB1C3 |
| Submitted by | [http://2016.igem.org/Team:NEU-China] |
The Freiburg iGEM team 2013 designed a fusion protein consisting of dCas9 and VP16 [1-6] for sequence-specific transactivation of a desired target locus [http://2013.igem.org/Team:Freiburg/Project/effector#activation (more information)]. Therefore, we used our double mutated dCas9 (BBa_K1150000) impaired in its cleavage activity and fused it to the 5’ end of the sequence coding for the transactivation domain of VP16 (BBa_K1150001). To ensure nuclear localization of the construct a nuclear localization signal (NLS) was fused to both ends of dCas9-VP16. For detection of expression the fusion protein was tagged with a HA-epitope coding sequence (BBaa_K1150016) and its expression was set under control of the CMV promoter (BBa_K747096) and BGH terminator (BBa_K1150012). Figure 1 illustrates the detailed design of the whole device.
Figure 1: Construct design. dCas9 was fused via a 3 amino acid linker to VP16. The resulting fusion construct was flanked by NLS sequences and tagged by a HA epitope. The CMV promoter and BGH terminator were chosen to control gene expression.
By co-transfecting our RNA plasmid (BBa_K1150034) which includes the tracrRNA and the separately integrated, desired crRNA, the dCas9 specifically binds to the targeted DNA sequence. With the help of the transactivation domain of VP16, transcription factors are recruited and the pre-initiation complex can be built. By placing this construct upstream of a promotor region any gene of interest can be activated.
Figure 2: Principle of transactivation of mammalian gene expression by the fusion protein dCas9-VP19 (BBa_K1150020). The double mutated dCas9 (D10A; H840A) fused to the herpes simplex virus (HSV) derived VP16 activation domain can serve as a crRNA-guided DNA-binding and transactivating protein. If a PAM sequence is present at the 3’ end of the crRNA binding site almost any DNA sequence can be targeted. Abbr.: Pmin: minimal promoter, containing minimal requirements for binding of transcription factors; Cas9: CRISPR associated protein 9; crRNA: CRISPR RNA; tracrRNA: trans-activating RNA; VP16: Virus protein 16, herpes simplex virus (HSV)-derived transcriptional activator protein; PAM: protospacer adjacent motif.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 4444
Illegal BamHI site found at 1214
Illegal BamHI site found at 3313 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1049
Illegal AgeI site found at 3148 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1031
Illegal SapI site found at 3130