Difference between revisions of "Part:BBa K864401"
(→NJTech_China 2020’s Characterisation) |
(→NJTech_China 2021’s Characterisation) |
||
| Line 36: | Line 36: | ||
Results | Results | ||
<p> </p> | <p> </p> | ||
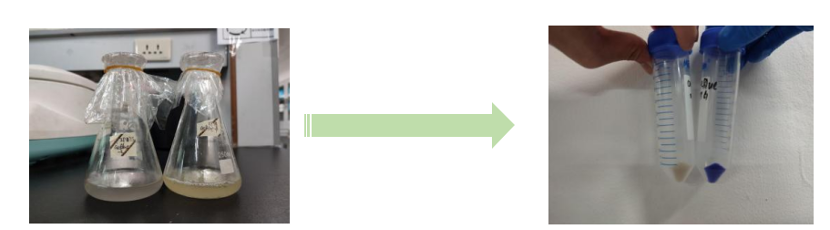
| − | [[File:NJTech_China_aeBlue-1.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.1</b> The fermentation broth of aeBlue.   <b>Fig.2</b> The cell pellet collected after centrifugation.]] | + | [[File:NJTech_China_aeBlue-1.png|width='100%' valign='top'| |center|thumb|550px|''<b>Fig.1</b> The fermentation broth of aeBlue.    <b>Fig.2</b> The cell pellet collected after centrifugation.]] |
<br> | <br> | ||
Conclusion: The cell pellet was collected by harvesting 50mL culture after 24h of induction followed by centrifugation at 4 degrees and 6000 rpm for 10min. Then, we performed ultrasonic disruption and collected the supernatant after centrifugation. The protein was purified and collected through ultrafiltration and affinity chromatography. | Conclusion: The cell pellet was collected by harvesting 50mL culture after 24h of induction followed by centrifugation at 4 degrees and 6000 rpm for 10min. Then, we performed ultrasonic disruption and collected the supernatant after centrifugation. The protein was purified and collected through ultrafiltration and affinity chromatography. | ||
Revision as of 07:24, 2 October 2021
aeBlue blue chromoprotein
aeBlue is a blue chromoprotein extracted from the basal disk of a beadlet anemone Actinia equina. The protein has an absorption maximum at 597nm and a deep blue colour. The protein aeBlue has significant sequence homologies with proteins in the GFP family.
Important: This part is not available in the registry yet, however, the same part is available from the registry with the standard RBS B0034: BBa_K1033929.
Source
The protein was first extracted and characterized by Shkrob et. al. 2005 under the name aeCP597. This version is codon optimized for E coli by Bioneer Corp.
Below to the left is the expressed aeBlue in E coli. To the right comparison of aeBlue with other chromoproteins available from the registry.

iGEM12_Uppsala_University: Left picture: Visual color expression of aeBlue(BBa_K864401).
Right picture: The Uppsala chromoprotein collection and RFP. Pellets of bacteria expressing chromoproteins eforRed (BBa_K592012), RFP (BBa_E1010), cjBlue (BBa_K592011), aeBlue (BBa_K864401), amilGFP (BBa_K592010) and amilCP (BBa_K592009).
Characterization
NJTech_China 2021’s Characterisation
Group: NJTech_China iGEM 2021
While most of our project was focused on Design and Construction of Synthetic Yeast-Microalgae Consortia for Biosynthesis of Phenylethanol, we were also interested in chromoproteins. Specifically, we characterized the expression of aeBlue and gfasPurple.
The aeBlue sequence (Part:BBa_K864401) optimized for E. coli was incorporated into plasmid pET-29a(+), transformed into E. coli BL21 for characterization and measurement. We provided aeBlue with results and data based on protein expression and purification, TOF-Mass spectrometry, full wavelength measurement and Swiss-model.
Methods:
SDS-PAGE, TOF-Mass Spectrometry, BCA (Bicinchoninic acid) method, full wavelength measurement and Swiss-Model.
Results
Conclusion: The cell pellet was collected by harvesting 50mL culture after 24h of induction followed by centrifugation at 4 degrees and 6000 rpm for 10min. Then, we performed ultrasonic disruption and collected the supernatant after centrifugation. The protein was purified and collected through ultrafiltration and affinity chromatography.
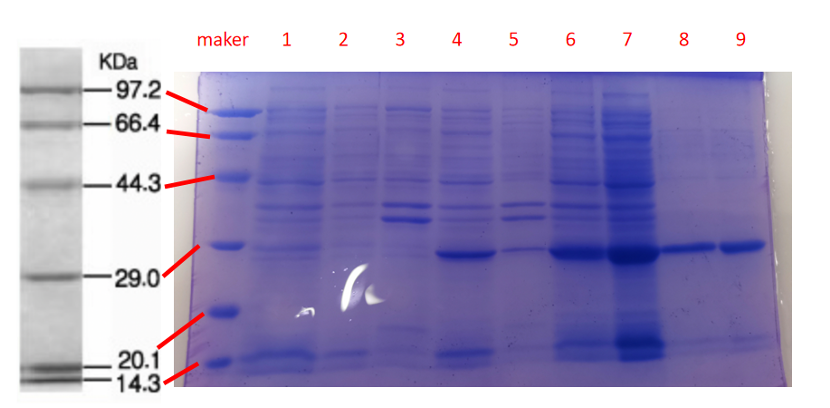
1· aeBlue- The culture after IPTG induction.
2· aeBlue- The pellet after IPTG induction and ultrasound.
3· aeBlue- Supernatant after IPTG induction and sonication.
4· aeBlue- The culture without IPTG induction.
5· aeBlue- The pellet without IPTG induction after ultrasound.
6· aeBlue- Supernatant sample without IPTG induction after sonication.
7· aeBlue- Protein sample after the ultrafiltration (diluted 20 times).
8· aeBlue- Purified protein sample.
Conclusion: The protein gel preliminarily proved that the molecular mass of the aeBlue protein was correct, which is consistent with the expected molecular mass of aeBlue protein (the molecular mass of aeBlue protein is about 27.3 kDa). Compared with lane 1, 2 and 3, lane 4, 5, and 6 indicate that more aeBlue protein can be obtained with IPTG induction. As is shown in lane 7, the concentration of protein was increased after ultrafiltration concentration. Lane 8 shows that the purification effect of protein after nickel affinity chromatography was better, and the impurity protein was less than before affinity chromatography. In conclusion, it can be seen that our expression and purification strategy is effective.
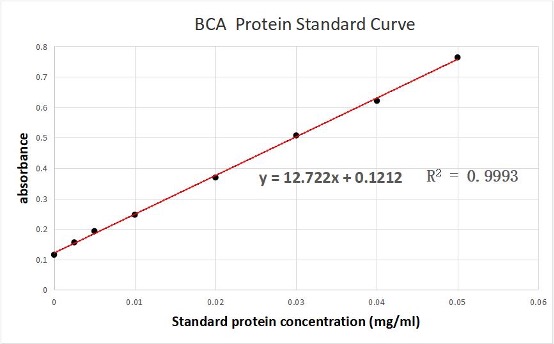
We used the BCA (Bicinchoninic acid) method to measure the concentration of aeBlue protein.
The concentration of aeblue chromoprotein was 9.10 mg/ml.
The standard protein curve fitting equation (R2=0.9993) :
y=12.722x+0.1212
It comes out that:
The concentration of aeBlue is 9.10 mg/ml.
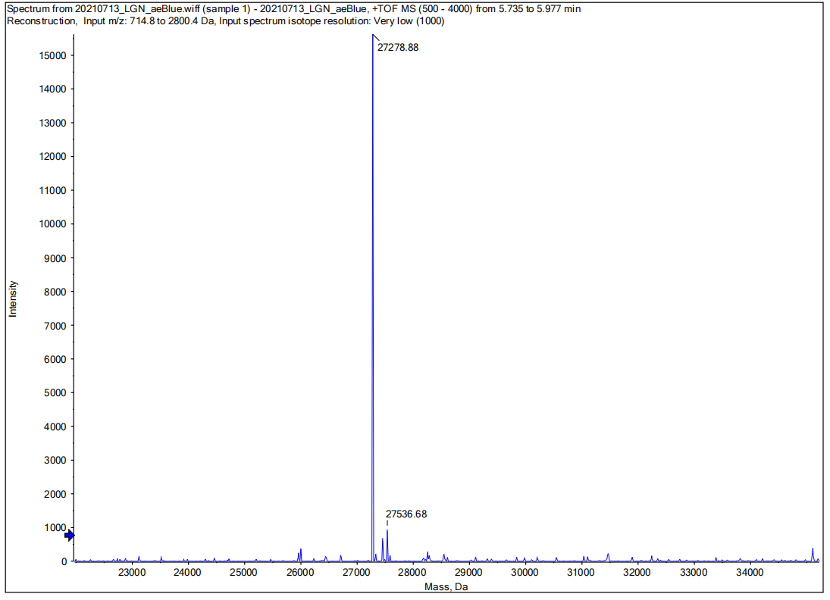
Conclusion : We performed Time of Flight Mass Spectrometer on the purified HIS-tagged aeBlue protein. The predicted molecular mass of this protein is about 27300Da. The result of TOF-Mass Spectrometry showed that the specific molecular mass of aeBlue protein is 27.279kDa (the value of the sharpest peak is shown as the molecular mass of aeBlue protein). Moreover, the intensity of 27.279kDa is up to 1.5×10^5, which indicates the high concentration and purity of the aeBlue protein. There are also some small protein peaks, suggesting that the noise had some effect, but not much.
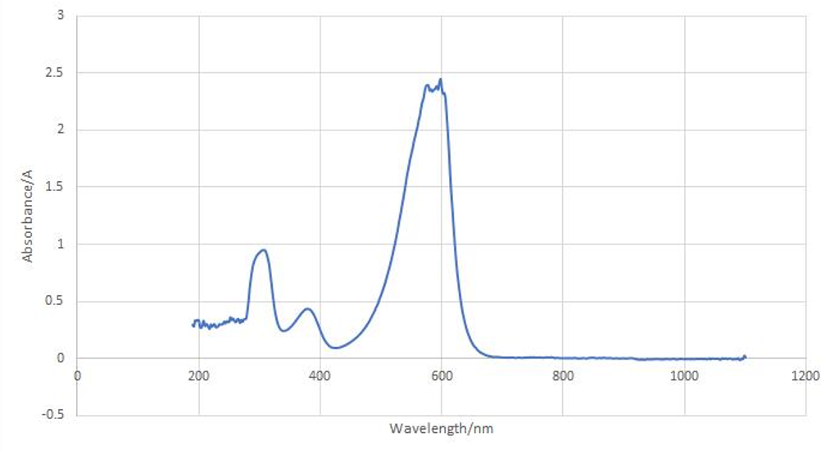
aeBlue protein full-wavelength scan profile :
1-204nm 0.282A
2-598nm 2.446A
3-272nm 0.322A
4-1040nm 0.003A
Conclusion : The full-wavelength scan of aeBlue protein shows that the strongest absorption peak of aeBlue protein occurs at 598nm. As shown in the results, aeBlue has a low intensity peak at 204 to 272 nm, which may be due to the fluorescence excitation.
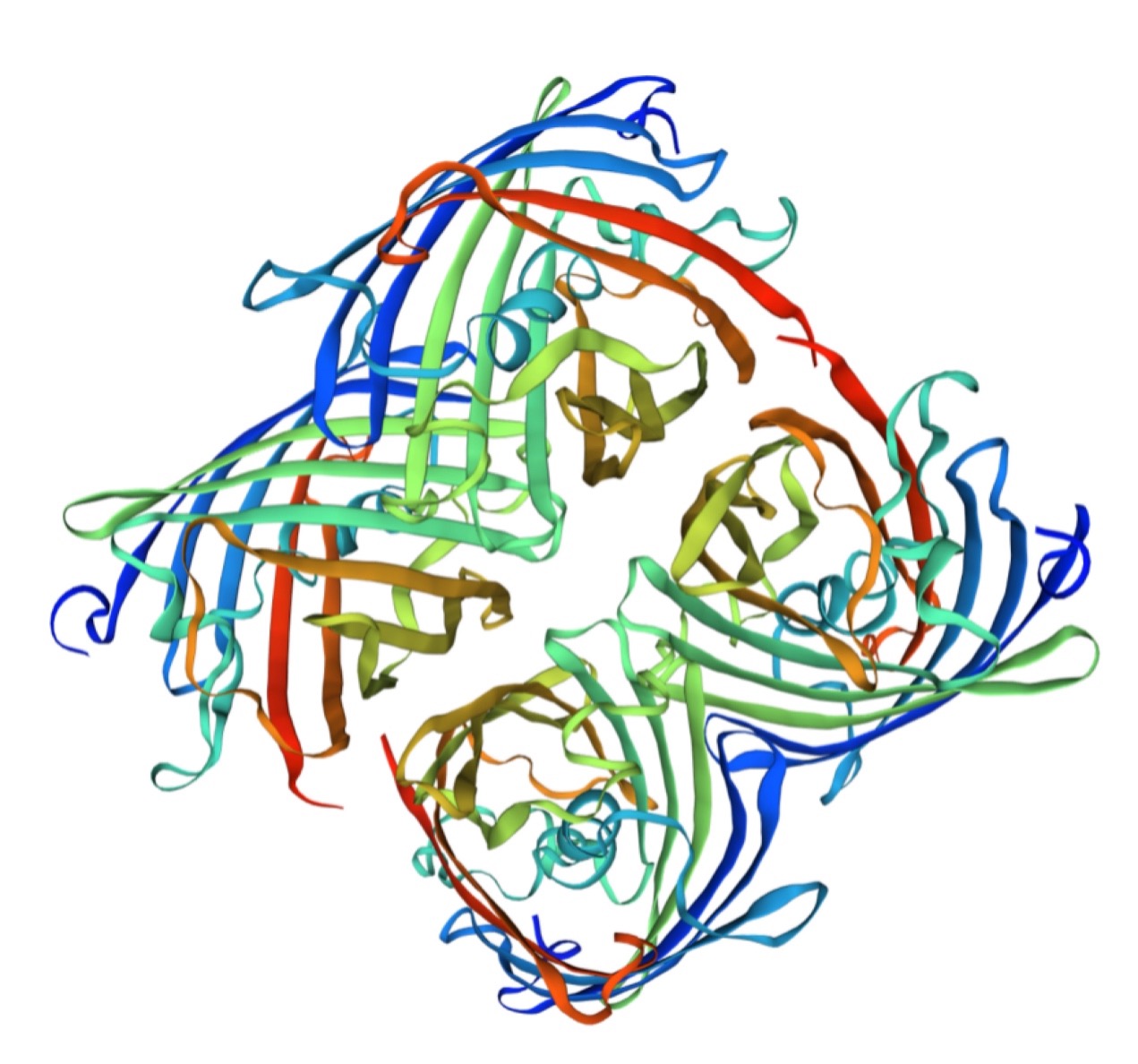
Structural modeling results of the aeBlue protein based on Swiss-Model
Conclusion: We used Swiss-Model to simulate the three-dimensional structure of aeBlue protein. The above figures showed the modeling result of Swiss-Model.
References
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1316306/]Shkrob, M.A., Yanushevich, Y.G., Chudakov, D.M., Gurskaya, N.G., Labas, Y.A., Poponov, S.Y., Mudrik, N.N., Lukyanov, S., Lukyanov, K.A., 2005. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem. J. 392, 649–654.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]