Difference between revisions of "Help:Plasmid backbones/Features"
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Help:Plasmid backbones|< Back to Plasmid backbone Help]] | |
| − | + | [[Image:BioBrickvectorbackbone.png|right]] | |
| − | + | <font size="4">'''Plasmid backbones tend to have three key features'''</font> | |
| − | + | ||
| − | + | ||
| − | <font size="4">''' | + | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | = | + | <font size="4">1. Replication origin</font> |
| − | + | ---- | |
| − | + | {{:Help:Plasmid backbones/Features/Replication_origin}} | |
| − | + | ||
| + | Thus, you often [[Help:Plasmid backbones/Choosing the right plasmid|choose your plasmid backbone replication origin]] based on your application. | ||
| + | |||
| + | |||
| + | <font size="4">2. Antibiotic resistance marker</font> | ||
| + | ---- | ||
| + | {{:Help:Plasmid backbones/Features/Antibiotic_resistance_marker}} | ||
| + | |||
| + | |||
| + | <font size="4">3. Cloning site</font> | ||
| + | ---- | ||
| + | The '''cloning site''' is the location on the plasmid backbone where BioBrick parts are inserted. Most plasmid backbones in microbiology have "multiple cloning site" because the plasmid backbone has several restriction sites in a row allowing you to choose where you insert your DNA fragment and which enzymes you use. In contrast, BioBrick plasmid backbones have a BioBrick cloning site consisting of a BioBrick prefix (including enzyme recognition sites for EcoRI and XbaI) and a BioBrick suffix (including enzyme recognition sites for SpeI and PstI). All BioBrick parts are inserted at the BioBrick cloning site between the BioBrick prefix and BioBrick suffix. The prefix sequence occurs ''before'' the BioBrick part, and the suffix sequence occurs ''after'' the BioBrick part. | ||
| + | |||
| + | __NOTOC__ __NOEDITSECTION__ | ||
Latest revision as of 18:51, 10 December 2008
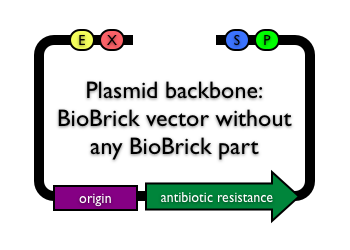
< Back to Plasmid backbone Help
Plasmid backbones tend to have three key features
1. Replication origin
The genetic element responsible for the replication of plasmids during cell growth and division is called a replication origin (also "origin of replication" or simply "origin"). There are several different replication origins and they differ in their plasmid copy number per cell (how many molecules of the plasmid are maintained in the cell), mechanism of copy number control, cell-to-cell copy number variation, and even the degree of coiling of the physical DNA. Thus, BioBrick® parts, devices and systems can operate very differently from one plasmid backbone to another.
Thus, you often choose your plasmid backbone replication origin based on your application.
2. Antibiotic resistance marker
At the most basic level, function of the antibiotic resistance marker is to allow the cell to grow even in the presence of a particular antibiotic. Plasmid backbones include antibiotic resistance markers because the markers allow you to select for cells that contain your plasmid. When E. coli cells grow and divide, plasmids can inadvertently be lost from the cell. In some cases, cells without a plasmid can potentially grow faster than cells with the plasmid which means that cell cultures can quickly become dominated by plasmid-free cells. Since most BioBrick® parts are maintained on plasmid backbones, plasmid loss is problematic. To help avoid these problems, every plasmid backbone includes an antibiotic resistance marker. Thus, cells which don't have a copy of the plasmid are killed by antibiotic present. Common antibiotic resistance markers in BioBrick® plasmid backbones confer resistance to ampicillin ("Amp" or A), kanamycin ("Kan" or K), chloramphenicol ("Cm" or C) and tetracycline ("Tet" or T).
3. Cloning site
The cloning site is the location on the plasmid backbone where BioBrick parts are inserted. Most plasmid backbones in microbiology have "multiple cloning site" because the plasmid backbone has several restriction sites in a row allowing you to choose where you insert your DNA fragment and which enzymes you use. In contrast, BioBrick plasmid backbones have a BioBrick cloning site consisting of a BioBrick prefix (including enzyme recognition sites for EcoRI and XbaI) and a BioBrick suffix (including enzyme recognition sites for SpeI and PstI). All BioBrick parts are inserted at the BioBrick cloning site between the BioBrick prefix and BioBrick suffix. The prefix sequence occurs before the BioBrick part, and the suffix sequence occurs after the BioBrick part.