|
|
| Line 1: |
Line 1: |
| | [[DNA|< Back to DNA parts]] | | [[DNA|< Back to DNA parts]] |
| − |
| |
| − | <center>
| |
| − | [[DNA/Recombination#Salmonella typhimurium-derived Hin/hix DNA recombination system|'''''Salmonella'' Hin/''hix''''']] • [[DNA/Recombination#Bacteriophage P1 Cre/lox DNA recombination system|'''P1 Cre/''lox''''']] • [[DNA/Recombination#Bacteriophage λ att DNA recombination system|'''λ ''att''''']] • [[DNA/Recombination#Bacteriophage P22 att DNA recombination system|'''P22 ''att''''']] • [[DNA/Recombination#Saccharomyces cerevisiae-derived Flp/FRT DNA recombination system|'''Yeast Flp/''FRT''''']] • [[DNA/Recombination#E. coli XerCD/dif DNA recombination system|'''''E. coli'' XerCD/''dif''''']]
| |
| − | </center>
| |
| − |
| |
| − | <html>
| |
| − | <style>
| |
| − | #assembly_plasmid_table {margin-left:auto;margin-right:auto; border: 3px solid #444444;}
| |
| − | #assembly_plasmid_table td {background-color:#eeeeee; padding: 5px; font-size: 12px;}
| |
| − | #assembly_plasmid_table th {background-color: #aaaaaa;padding: 5px; font-size:14px;}
| |
| − | </style>
| |
| − | </html>
| |
| | | | |
| | {{:DNA/Recombination/Overview}} | | {{:DNA/Recombination/Overview}} |
| | | | |
| − | Site-specific recombination systems derived from ''Salmonella'', different bacteriophages, yeast, and ''E. coli'' are all available from the Registry. For more details, see below. | + | Site-specific recombination systems derived from ''Salmonella'', different bacteriophages, yeast, and ''E. coli'' are all available from the Registry. For more details on individual DNA recombination systems, click the links below. |
| − | | + | |
| − | ==''Salmonella typhimurium''-derived Hin/''hix'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | |[[Image:KarmellaHaynesPhoto.jpg|60px|center]]
| + | |
| − | |Karmella Haynes and the [http://parts.mit.edu/wiki/index.php/Davidson_2006 2006 Davidson College/Missouri Western iGEM team], designed and constructed a set of parts from the ''Salmonella typhimurium''-derived DNA recombination system. You can read more about the 2006 Davidson/Missouri Western project in their open-access paper [http://www.jbioleng.org/content/2/1/8 Engineering bacteria to solve the Burnt Pancake Problem] published in the ''Journal of Biological Engineering'' <cite>Haynes</cite>. The following is excerpted from their paper.
| + | |
| − | |}
| + | |
| − | | + | |
| − | In ''Salmonella'', Hin DNA recombinase ([[Part:BBa_J31000|BBa_J31000]], [[Part:BBa_J31001|BBa_J31001]]) catalyzes an inversion reaction that regulates the expression of alternative flagellin genes by switching the orientation of a promoter located on a 1 kb invertible DNA segment <cite>Zieg,Zieg80</cite>. Two palindromic 26 bp ''hix'' sequences flank the invertible DNA segment and serve as the recognition sites for cleavage and strand exchange. A ~70 bp cis-acting ''recombinational enhancer (RE)'' increases efficiency of protein-DNA complex formation ([[Part:BBa_J3101|BBa_J3101]]) <cite>Johnson</cite>. We have reconstituted the genetic elements required for DNA inversion as a collection of modular genetic elements for use in ''E. coli''. Our system is a proof-of-concept genetic computing device that manipulates plasmid DNA processors within living cells.
| + | |
| − | | + | |
| − | In ''Salmonella'', the asymmetrical palindromic sequences ''hixL'' and ''hixR'' flank the invertible DNA segment and serve as the recognition sites for cleavage and strand exchange. Our system uses ''hixC'' ([[Part:BBa_J44000|BBa_J44000]]), a composite symmetrical hix site that shows higher binding affinity for Hin and a 16-fold slower inversion rate than wild type sites ''hixL'' and ''hixR'' <cite>Lim,Moskowitz</cite>.
| + | |
| − | | + | |
| − | We have demonstrated that a modified Hin/''hix'' DNA recombination system can be used ''in vivo'' to manipulate at least two adjacent ''hixC''-flanked DNA segments; HinLVA and ''hixC'' are sufficient for DNA inversion activity. The Hin/''hix'' DNA recombination system could be used for other biological engineering applications. We have developed a set of modular genetic elements, ''hixC'' ([[Part:BBa_J44000|BBa_J44000]]), ''RE'' ([[Part:BBa_J3101|BBa_J3101]]), and ''HinLVA'' ([[Part:BBa_J31001|BBa_J31001]]), that expands the repertoire of molecular tools for enzyme-mediated DNA manipulation ''in vivo''.
| + | |
| − | | + | |
| − | <parttable>hix_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/hix" under the Hard Information tab of the part. -->
| + | |
| − | | + | |
| − | ===References===
| + | |
| − | <biblio>
| + | |
| − | #Zieg pmid=322276
| + | |
| − | #Zieg80 pmid=6933466
| + | |
| − | #Johnson pmid=2548848
| + | |
| − | #Haykinson pmid=8508775
| + | |
| − | #PerkinsBalding pmid=9244261
| + | |
| − | #Haynes pmid=18492232
| + | |
| − | #Ham pmid=18665232
| + | |
| − | #Nanassy pmid=9691026
| + | |
| − | #Moswitz pmid=1885005
| + | |
| − | #Lim pmid=1597453
| + | |
| − | </biblio>
| + | |
| − | | + | |
| − | | + | |
| − | ==Bacteriophage P1 Cre/''lox'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | |[[Image:JCA Photo.png|50px|center]]
| + | |
| − | |Chris Anderson, a professor of bioengineering at UC Berkeley, constructed the ''lox'' recombination site [[Part:BBa_J61046|BBa_J61046]].
| + | |
| − | |[[Image:NoPhotoAvailable.jpg|60px|center]]
| + | |
| − | |Eimad Shotar, a member of the [http://parts.mit.edu/igem07/index.php/Paris 2007 Paris iGEM team], constructed the ''lox'' recombination sites [[Part:BBa_I718016|BBa_I718016]] and [[Part:BBa_I718017|BBa_I718017]].
| + | |
| − | |}
| + | |
| − | | + | |
| − | ''The following text is excerpted from Siegel ''et al.'' <cite>Siegel04</cite>.''
| + | |
| − | | + | |
| − | Bacteriophage P1 uses a site-specific recombination system that is responsible for partitioning newly synthesized genomic copies during replication <cite>Abremski, Hoess</cite>. This system is composed of a 38-kD phage-encoded Cre recombinase that mediates symmetrical recombination between two 34-bp ''loxP'' sites <cite>Abremski</cite>, which are recreated after recombination. Recombination between two compatible ''loxP'' sites will excise or invert the intervening DNA in the case of an intramolecular reaction or transfer suitably flanked ''loxP'' DNA in an intermolecular double cross-over recombination event. The Cre/''loxP'' system does not require accessory factors to carry out recombination ''in vivo'' or ''in vitro'', and studies have identified several hetero-specific ''loxP'' sequences that exclusively recombine with themselves, but not with wild-type ''lox'' <cite>Hoess86, Sauer92, Lee98, Siegel01</cite>. Importantly, the Cre/''lox'' system has also been shown to be functional in site-specific recombination in mammalian cell lines <cite>Sauer88</cite>.
| + | |
| − | | + | |
| − | <parttable>lox_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/lox" under the Hard Information tab of the part. -->
| + | |
| − | | + | |
| − | ===References===
| + | |
| − | <biblio>
| + | |
| − | #Abremski pmid=6319400
| + | |
| − | #Hoess pmid=6230671
| + | |
| − | #Hamilton pmid=6333513
| + | |
| − | #Hoess86 pmid=3457367
| + | |
| − | #Sauer88 pmid=2839833
| + | |
| − | #Sauer92 pmid=1554399
| + | |
| − | #Lee98 pmid=9714735
| + | |
| − | #Sauer98 pmid=9608509
| + | |
| − | #Siegel01 pmid=11576551
| + | |
| − | #Sauer02 pmid=12624421
| + | |
| − | #Siegel04 pmid=15173117
| + | |
| − | </biblio>
| + | |
| − | | + | |
| − | | + | |
| − | ==Bacteriophage λ ''att'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | |[[Image:NoPhotoAvailable.jpg|60px|center]]
| + | |
| − | |The 2004 Boston University iGEM team designed and constructed the ''att'' recombination sites [[Part:BBa_I11022|BBa_I11022]] and [[Part:BBa_I11023|BBa_I11023]].
| + | |
| − | |}
| + | |
| − | | + | |
| − | ''The following is excerpted from Radman-Livaja et al. <cite>RadmanLivaja</cite> and Landy et al. <cite>Landy</cite>. It has been edited for clarity.''
| + | |
| − | | + | |
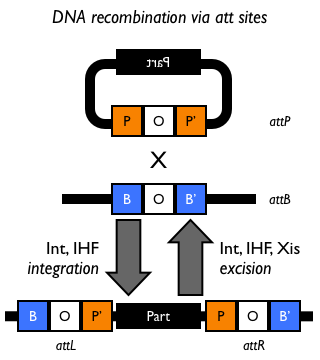
| − | Bacteriophage λ has long served as a model system for studies of regulated site-specific recombination. In conditions favorable for bacterial growth, the phage genome is inserted into the ''Escherichia coli'' genome by an ‘integrative’ recombination reaction, which takes place between DNA attachment sites called ''attP'' and ''attB'' in the phage and bacterial genomes, respectively. As a result, the integrated λ DNA is bounded by hybrid attachment sites, termed ''attL'' and ''attR''. In response to the physiological state of the bacterial host or to DNA damage, λ phage DNA excises itself from the host chromosome. This excision reaction recombines attL with attR to precisely restore the ''attP'' and ''attB'' sites on the circular λ and ''E. coli'' DNAs <cite>Campbell</cite>. The phage-encoded λ integrase protein (Int), a tyrosine recombinase, splices together bacterial and phage attachment sites. Int is required for both integration and excision of the λ prophage <cite>Zissler67</cite>.
| + | |
| − | | + | |
| − | λ recombination has a strong directional bias in response to environmental conditions. Accessory factors, whose expression levels change in response to host physiology, control the action of Int and determine whether the phage genome will remain integrated or be excised. Int has two DNA-binding domains: a C-terminal domain, consisting of a catalytic domain and a core-binding (CB) domain, that interacts with the core recombining sites and an N-terminal domain (N-domain) that recognizes the regulatory arm DNA sites <cite>Wojciak02</cite>. The heterobivalent Int molecules bridge distant core and arm sites with the help of accessory proteins, such as integration host factor (IHF), which bend the DNA at intervening sites, and appose arm and core sequences for interaction with the Int recombinase. Five arm DNA sites in the regions flanking the core of ''attP'' are differentially occupied during integration and excision reactions. The integration products ''attL'' and ''attR'' cannot revert back to ''attP'' and ''attB'' without assistance from the phage-encoded factor Xis, which bends DNA on its own or in combination with the host-encoded factor Fis <cite>Abremski82,Abremski81,Hoess80,Thompson87a,Thompson87b,Ball91</cite>. Xis also inhibits integration, and prevents the ''attP'' and ''attB'' products of excision from reverting to ''attL'' and ''attR'' <cite>Abremski81,18]. Excision is inhibited by high concentrations of IHF <cite>Bushman85,Thompson86</cite>. Because the cellular levels of IHF and Fis proteins respond to growth conditions, these host-encoded factors have been proposed as the master signals for integration and excision <cite>Thompson87a,Bushman85,Thompson86, Nilsson92,Ball92, Ball91</cite>.
| + | |
| − | | + | |
| − | ''The following is excerpted from Landy et al. <cite>Landy</cite>. It has been edited for clarity.''
| + | |
| − | | + | |
| − | The phage (''attP'') and bacterial (''attB'') ''att'' sites are designated POP’ and BOB’, respectively, and the prophage ''att'' sites are designated BOP’ (''attL'') and POB’ (''attR''). Transducing phage carrying ''attL'' (λ''gal'') or ''attR'' (λ''bio'') are generated from a prophage which has excised from the host chromosome by a rare ''int''-independent recombination which deletes phage DNA from one end of the prophage and adds bacterial DNA to the other <cite>Campbell</cite>. A phage carrying the bacterial ''att'' site, BOB’, is obtained as a product of ''int''-promoted recombination between a ''gal'' (BOP’) and a ''bio'' (POB’) transducing phage which is capable of transducing gal and bio together <cite>Echols70</cite>.
| + | |
| − | | + | |
| − | When ''Escherichia coli'' carries a deletion of the primary bacterial ''att'' site BOB’, ''int''-dependent integration of λ can be detected at numerous loci (secondary bacterial ''att'' sites) on the ''E. coli'' chromosome <cite>Shimada72</cite>. This integration always involves the phage att site (POP’) <cite>Shimada75</cite> and is thus very similar to the behavior of IS-elements <cite>Fiandt72, Hirsch72</cite>. The secondary prophage att sites are given the general designation ΔOP’ and POΔ’ and they differ from BOP’ and POB’ and from each other in their biological properties as determined by ''int''- and ''xis'' -dependent recombination frequencies with various ''att'' sites.
| + | |
| − | | + | |
| − | <parttable>lambda_att_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/lambdaatt" under the Hard Information tab of the part. -->
| + | |
| − | | + | |
| − | ===References===
| + | |
| − | <biblio>
| + | |
| − | #Campbell Campbell, AM. Episomes. In: Caspari EW, Thoday JM. , editors. Advances in Genetics. 1. New York: Academic Press; 1962. pp. 101–145.
| + | |
| − | #Zissler67 pmid=5637199
| + | |
| − | #Guameros70 pmid=4907272
| + | |
| − | #Kaiser70 pmid=4907271
| + | |
| − | #Echols70 pmid=4907273
| + | |
| − | #Shimada72 pmid=4552408
| + | |
| − | #Shimada75 pmid=1095763
| + | |
| − | #Fiandt72 pmid=4567155
| + | |
| − | #Hirsch72 pmid=4567154
| + | |
| − | #Hoess80 pmid=6446713
| + | |
| − | #Abremski81 pmid=6279866
| + | |
| − | #Abremski82 pmid=6213611
| + | |
| − | #Bushman85 pmid=2932798
| + | |
| − | #Thompson86 pmid=2946666
| + | |
| − | #Thompson87a pmid=2957063
| + | |
| − | #Thompson87b pmid=2958633
| + | |
| − | #Kitts88a pmid=2970060
| + | |
| − | #Kitts88b pmid=2975338
| + | |
| − | #NunesDuby87 pmid=3040260
| + | |
| − | #Holliday64 Holliday, R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304.
| + | |
| − | #Ball91 pmid=1829453
| + | |
| − | #Ball92 pmid=1459953
| + | |
| − | #Nilsson92 pmid=1732224
| + | |
| − | #Hallett97 pmid=9348666
| + | |
| − | #Azaro02 Azaro, MA; Landy, A. λ Int and the λ Int family. In: Craig NL, Craigie R, Gellert M, Lambowitz A. , editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148.
| + | |
| − | #Wojciak02 pmid=11904406
| + | |
| − | #Landy pmid=331474
| + | |
| − | #Landy89 pmid=2528323
| + | |
| − | #RadmanLivaja pmid=16368232
| + | |
| − | </biblio>
| + | |
| − | | + | |
| − | | + | |
| − | ==Bacteriophage P22 ''att'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | |[[Image:NoPhotoAvailable.jpg|60px|center]]
| + | |
| − | |The 2004 Boston University iGEM team designed and constructed the P22 ''att'' recombination sites [[Part:BBa_I11032|BBa_I11032]] and [[Part:BBa_I11033|BBa_I11033]].
| + | |
| − | |}
| + | |
| − | | + | |
| − | ''The following text is excerpted from Cho et al. <cite>Cho99</cite>. It has been edited for clarity.''
| + | |
| − | | + | |
| − | Bacteriophage P22 is a lambdoid phage which infects ''Salmonella typhimurium''. P22 can integrate into and excise out of its host chromosome via site-specific recombination. Both integration and excision reactions require the phage-encoded ''int'' gene <cite>Smith67</cite>, and excision is dependent on the ''xis'' gene as well.
| + | |
| − | | + | |
| − | P22 Int is a member of the λ integrase family <cite>Argos86, Esposito97, NunesDuby98</cite>. The Int proteins of λ and P22 are composed of two domains. The catalytic domain binds to the core region of the phage recombination site, ''attP'', where the actual recombination reactions occur. The smaller amino-terminal domain binds to arm-type sequences which are located on either site of the core within the attP <cite>Vargas88, Mungo94</cite>. The active components of λ integrative and excisive recombination are nucleosome-like structures, called intasomes, in which DNA is folded around several molecules of Int and integration host factor (IHF) <cite>Better82, Griffith85, Pollock83, Richet88, Robertson88</cite>. It has been demonstrated that one monomer of λ integrase can simultaneously occupy both a core-type binding site and an arm-type binding site <cite>Kim90, MacWilliams96</cite>. Formation of these bridges is facilitated by IHF, which binds to specific sequences and imparts a substantial bend to the DNA <cite>Craig84, Rice96, Robertson88, Snyder89</cite>.
| + | |
| − | | + | |
| − | The ''attP'' regions of P22 and λ are also similar in that both contain arm regions, known as the P and P′ arms, which contain Int arm-type binding sites and IHF binding sites <cite>Leong86, Mungo94</cite>. However, the arrangement, spacing, and orientation of the Int and IHF binding sites are distinct <cite>Mungo94</cite>. The ''attP'' region of λ contains two Int arm-type binding sites on the P arm and three on the P′ arm. The P arm contains two IHF binding sites, and the P′ arm contains a single site. The ''attP'' region of P22 contains three Int arm-type binding sites on the P arm and two sites on the P′ arm. In addition, IHF binding sites, called H and H′, are located on each arm of the P22 ''attP''. Leong ''et al.'' <cite>Leong85</cite> showed that the ''Escherichia coli'' IHF can recognize and bind to these P22 IHF binding sites ''in vitro''. It was also shown that the maximum amount of P22 integrative recombination occurred in the presence of ''E. coli'' IHF ''in vitro'', whereas in its absence, recombination was detectable but depressed <cite>Mungo94</cite>. However, the requirement for IHF or other possible accessory proteins during P22 site-specific recombination ''in vivo'' has not been tested. In this study, we assessed the role of IHF in P22 integration and excision ''in vivo''.
| + | |
| − | | + | |
| − | Although the ''attP'' region of P22 contains strong IHF binding sites, ''in vivo'' measurements of integration and excision frequencies showed that infecting P22 phages can perform site-specific recombination to its maximum efficiency in the absence of IHF. In addition, a plasmid integration assay showed that integrative recombination occurs equally well in wild-type and ''ihfA'' mutant cells. P22 integrative recombination is also efficient in Escherichia coli in the absence of functional IHF. These results suggest that nucleoprotein structures proficient for recombination can form in the absence of IHF or that another factor(s) can substitute for IHF in the formation of complexes.
| + | |
| − | | + | |
| − | <parttable>P22_att_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/P22att" under the Hard Information tab of the part. -->
| + | |
| − | | + | |
| − | ===References===
| + | |
| − | <biblio>
| + | |
| − | #Cho99 pmid=10400581
| + | |
| − | #Smith67 Smith, H O; Levine, M. A phage P22 gene controlling integration of prophage. Virology. 1967;31:297–316.
| + | |
| − | #Susskind78 pmid=353481
| + | |
| − | #Leong86 pmid=3491212
| + | |
| − | #Argos86 pmid=3011407
| + | |
| − | #Esposito97 pmid=9278480
| + | |
| − | #NunesDuby98 pmid=9421491
| + | |
| − | #Vargas88 pmid=2843292
| + | |
| − | #Mungo94 pmid=8051182
| + | |
| − | #Better82 pmid=6310548
| + | |
| − | #Griffith85 pmid=3159013
| + | |
| − | #Pollock83 pmid=6226803
| + | |
| − | #Richet88 pmid=2964274
| + | |
| − | #Robertson88 pmid=2831189
| + | |
| − | #Kim90 pmid=2146029
| + | |
| − | #MacWilliams96 pmid=8807282
| + | |
| − | #Craig84 pmid=6096022
| + | |
| − | #Rice96 pmid=8980235
| + | |
| − | #Snyder89 pmid=2528698
| + | |
| − | #Leong85 pmid=2984205
| + | |
| − | </biblio>
| + | |
| − | | + | |
| − | ==''Saccharomyces cerevisiae''-derived Flp/''FRT'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | |[[Image:JCA Photo.png|50px|center]]
| + | |
| − | |Chris Anderson, a professor of bioengineering at UC Berkeley, constructed the ''FRT'' recombination site [[Part:BBa_J61020|BBa_J61020]].
| + | |
| − | |}
| + | |
| − | | + | |
| − | ''The following text is excerpted from Huang et al. <cite>Huang</cite>. It has been edited for clarity.''
| + | |
| − | | + | |
| − | The FLP system of the yeast 2mm plasmid is one of the most attractive for genomic manipulation because of its efficiency, simplicity, and demonstrated ''in vivo'' activity in a wide range of organisms. The Flp system has been used to construct specific genomic deletions and gene duplications, study gene function, promote chromosomal translocations, promote site-specific chromosome cleavage, and facilitate the construction of genomic libraries in organisms including bacteria, yeast, insects, plants, mice, and humans. Site-specific recombination catalyzed by the FLP recombinase occurs readily in bacterial cells.
| + | |
| − | | + | |
| − | The yeast FLP system has been studied intensively (7, 8, 22, 36). The only requirements for FLP recombination are the FLP protein and the FLP recombination target (FRT) sites on the DNA substrates. The minimal functional FRT site contains only 34 bp. The FLP protein can promote both inter- and intramolecular recombination.
| + | |
| − | | + | |
| − | <parttable>FRT_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/FRT" under the Hard Information tab of the part. -->
| + | |
| − | | + | |
| − | ===References===
| + | |
| − | <biblio>
| + | |
| − | #Schweizer pmid=12736528
| + | |
| − | #Huang pmid=9324255
| + | |
| − | #Cherepanov pmid=7789817
| + | |
| − | #Cox pmid=6308608
| + | |
| − | #Fiering pmid=8378321
| + | |
| − | #Golic pmid=2509077
| + | |
| − | #Morris pmid=1945877
| + | |
| − | #Sadowski pmid=7659779
| + | |
| − | #Senecoff pmid=2997780
| + | |
| − | </biblio>
| + | |
| − | | + | |
| − | ==''E. coli'' XerCD/''dif'' DNA recombination system==
| + | |
| − | | + | |
| − | {|
| + | |
| − | | [[Image:XiaonanWangPhoto.jpg|100px]]
| + | |
| − | | Xiaonan Wang, a member of the [http://parts.mit.edu/igem07/index.php/Edinburgh 2007 University of Edinburgh iGEM team], designed the ''dif'' recombination sites [[Part:BBa_I742101|BBa_I742101]] and [[Part:BBa_I742102|BBa_I742102]].
| + | |
| − | |}
| + | |
| − | | + | |
| − | ''The following text is excerpted from Ip et al. <cite>Ip03</cite>.''
| + | |
| − | | + | |
| − | The separation and segregation of newly replicated [''E. coli''] circular chromosomes can also be prevented by the formation of circular chromosome dimers, which can arise during crossing over by homologous recombination <cite>Blakely91; Clerget91; Kuempel91</cite>. In ''E. coli'', these dimers, which arise about once every six generations, are resolved to monomers by the action of the FtsK–XerCD–dif chromosome dimer resolution machinery <cite>Steiner98a, Steiner98b, Recchia99, Steiner99</cite>. Two site-specific recombinases of the tyrosine recombinase family, XerCD, act at a 28 bp recombination site, ''dif'', located in the replication terminus region of the ''E. coli'' chromosome to remove the crossover introduced by dimer formation, thereby converting dimers to monomers. A complete dimer resolution reaction during recombination at dif requires the action of the C-terminal domain of FtsK (FtsK<sub>C</sub>) <cite>Steiner99; Barre00</cite>. FtsK is a multifunctional protein whose N-terminal domain acts in cell division, while the C-terminal domain functions in chromosome segregation <cite>Liu98; Wang98; Yu98a, Yu98b</cite>. Therefore, FtsK is well suited to coordinate chromosome segregation and cell division. A purified protein, FtsK<sub>50C</sub>, containing a functional C-terminal domain, can translocate DNA in an ATP-dependent manner and activate Xer recombination at the recombination site ''dif'', thereby reconstituting ''in vitro'' the expected ''in vivo'' activities of the C-terminal domain of the complete FtsK protein <cite>Aussel02</cite>.
| + | |
| − | | + | |
| − | <parttable>dif_recombination_site_DNA</parttable>
| + | |
| − | | + | |
| − | <!-- To include a part in this table, include the categories "//DNA/recombinationsite/dif" under the Hard Information tab of the part. -->
| + | |
| | | | |
| − | ===References===
| + | *[[DNA/Recombination/Salmonella typhimurium-derived Hin-hix|'''''Salmonella typhimurium''-derived Hin/''hix'' DNA recombination system''']] |
| − | <biblio>
| + | *[[DNA/Recombination/Bacteriophage P1-derived Cre-lox|'''Bacteriophage P1-derived Cre/''lox'' DNA recombination system''']] |
| − | #Blakely91 pmid=1931824
| + | *[[DNA/Recombination/Bacteriophage lambda-derived att|'''Bacteriophage λ-derived ''att'' DNA recombination system''']] |
| − | #Clerget91 pmid=1931823
| + | *[[DNA/Recombination/Bacteriophage P22-derived att|'''Bacteriophage P22 ''att'' DNA recombination system''']] |
| − | #Kuempel91 pmid=1657123
| + | *[[DNA/Recombination/Saccharomyces cerevisiae-derived Flp-FRT|'''Yeast Flp/''FRT'' DNA recombination system''']] |
| − | #Steiner98a pmid=9484882
| + | *[[DNA/Recombination/Escherichia coli-derived XerCD-dif|'''''Escherichia coli'' XerCD/''dif'' DNA recombination system''']] |
| − | #Steiner98b pmid=9829936
| + | |
| − | #Liu98 pmid=9723927
| + | |
| − | #Wang98 pmid=9723913
| + | |
| − | #Yu98a pmid=9495771
| + | |
| − | #Yu98b pmid=9829960
| + | |
| − | #Steiner99 pmid=10027974
| + | |
| − | #Recchia99 pmid=10523315
| + | |
| − | #Barre00 pmid=11114887
| + | |
| − | #Aussel02 pmid=11832210
| + | |
| − | #Ip03 pmid=14633998
| + | |
| − | </biblio>
| + | |
| | | | |
| | __NOTOC__ | | __NOTOC__ |
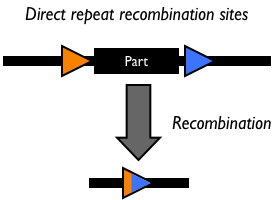
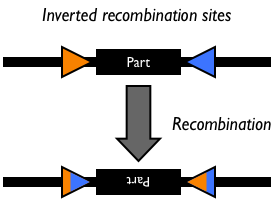
Site-specific DNA recombination requires both a recombinase protein and a pair of repeated DNA sites at which recombination takes place. Depending on the number and orientation of the DNA sites, there can be either an inversion, deletion or insertion of DNA (Figure 1). While some DNA recombination systems, such as Cre/lox only require the recombinase and the two DNA sites for recombination to occur, others either require or are modulated by additional accessory factors.