Difference between revisions of "Part:BBa B0010"
| (11 intermediate revisions by 8 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| + | {|width='80%' | ||
| + | |- | ||
| + | |width=35% valign='top'| | ||
| + | [[Image:Terminator.png]] | ||
| + | <partinfo>BBa_B0010 parameters</partinfo> | ||
| + | | width=50% valign='top' style='border: 1px solid black'| | ||
<partinfo>BBa_B0010 short</partinfo> | <partinfo>BBa_B0010 short</partinfo> | ||
| + | * Transcriptional terminator consisting of a 64 bp stem-loop. | ||
| − | + | |} | |
| + | |||
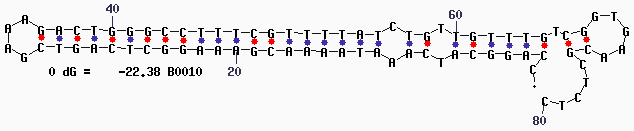
| + | '''Secondary Structure''' | ||
| + | |||
| + | [[Image:Mfold-B0010-1.png]] | ||
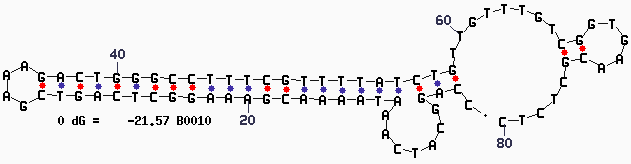
| + | |||
| + | [[Image:Mfold-B0010-2.png]] | ||
| + | |||
| + | |||
| + | {| width=50% style='border: 2px solid red' | ||
| + | |- | ||
| + | |'''Using primers VR/VF2 to PCR B0010 will result in excess bands.''' | ||
| + | '''See a full description of the problem [[Problems with PCR using VR/VF2|here]].''' | ||
| + | |- | ||
| + | |} | ||
| − | |||
| − | |||
| − | |||
<partinfo>BBa_B0010 SequenceAndFeatures</partinfo> | <partinfo>BBa_B0010 SequenceAndFeatures</partinfo> | ||
| − | + | <hr> | |
| + | '''Measurement''' | ||
| + | * [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured] | ||
| + | |||
| + | |||
| + | |||
| + | >Internal Priming Screening Characterization of BBa_B0010: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer. | ||
| + | |||
| + | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
<partinfo>BBa_B0010 parameters</partinfo> | <partinfo>BBa_B0010 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: -2.1 ± 3.4% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | ==Examining efficiency of ribosomal RNA operon B (rrnB) T1 terminator from E. coli.: NYU Abu Dhabi== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_B0010</partinfo> | ||
| + | |||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/7/79/NYUAD-BBa-B0010.png" style = "width:700px;"></center> | ||
| + | </div> | ||
| + | <figcaption><center>Figure: The intrinsic terminator is composed of the NusA protein, which interacts with the new RNA exiting out of the channel to stimulate termination. (Santangelo et. al., 2011).</center></figcaption> | ||
| + | </figure> | ||
| + | |||
| + | |||
| + | <p> Transcription of DNA is made up of the following steps: RNA polymerase binding to promoter and activation, initiation of RNA transcript, elongation of RNA transcript, and termination of transcription. Termination is the last step of transcription where the RNA polymerase releases the RNA transcript. Releasing the RNA transcript is not reversible and further transcription requires reinitiation at a promoter region to form a new RNA transcript (Uptain et. al, 1997). In prokaryotes like E. coli, the termination sites serve as targets for gene expression regulation as they can not only occur at the end of genes, but also near promoter regions or between genes in the operon (Nojima et. al., 2005). The T1 terminator region on the rrnB gene of E. coli is one of two terminator regions, the other being T2. The two termination regions have "factor-independent terminator-like sequences" with "two additional inverted repeats (IR1 and IR2) and a pair of direct repeats" (Orosz et. al., 1991). The T1 and T2 terminating regions are often used as an efficient terminator of transcription in many cloning vectors (Orosz et. al., 1991).</p> | ||
| + | |||
| + | <p>Efficiency is enhanced by the E. coli nusA protein, which gives effectiveness of inhibition in vitro comparable to those in vivo. These transcripts that are terminated when nusA protein is present are released from the RNA polymerase complex, suggesting that there is a complete termination reaction. The protein's termination factor activity is not dependent on the presence ofthe rho protein. The nusA protein serves as an antitermination factor, RNA polymerase subunit, and true termination factor at some terminator sites. In general, termination at T1 in vitro is quiteefficient with an 80% effectiveness rate without any additional factors. In vivo and in E. coliextracts, the T1 terminator has shown to be nearly 100% efficient. In an isolated and purified system with only nusA protein present, termination at this high level of efficiency is also achieved, suggesting that in vivo, the entity responsible for the highly efficient termination is dueto the nusA protein.</p> | ||
| + | |||
| + | <p>Literature used:</p> | ||
| + | <ul> | ||
| + | <li>Nojima, T., A. C. Lin, T. Fujii and I. Endo (2005). "Determination of the Termination Efficiencyof the Transcription Terminator Using Different Fluorescent Profiles in Green Fluorescent Protein Mutants." Analytical Sciences 21(12): 1479-1481.Nojima, T.; Lin, A. C.; Fujii, T.; Endo,I., Determination of the Termination Efficiency of the Transcription Terminator Using Different Fluorescent Profiles in Green Fluorescent Protein Mutants. Analytical Sciences 2005, 21 (12), 1479-1481.</li> | ||
| + | <li>Orosz, A., I. BOROS and P. VENETIANER (1991). "Analysis of the complex transcription termination region of the Escherichia coli rrn B gene." European journal of biochemistry 201(3): 653-659.</li> | ||
| + | <li>Schmidt, M. C. and M. J. Chamberlin (1987). "nusA Protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites." Journal of Molecular Biology 195(4): 809-818.</li> | ||
| + | <li>Uptain, S. M., & Chamberlin, M. J. (1997). Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proceedings of the National Academy of Sciences of the United States of America, 94(25), 13548–13553.</li> | ||
| + | <li>Santangelo, T. J. and I. Artsimovitch (2011). "Termination and antitermination: RNA polymerase runs a stop sign." Nature reviews. Microbiology 9(5): 319-329.</li> | ||
| + | </ul> | ||
| + | |||
| + | <p>This contribution was added by the <a href="https://2020.igem.org/Team:NYU_Abu_Dhabi/Contribution">2020 NYU Abu Dhabi team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 19:43, 27 October 2020
|
T1 from E. coli rrnB
|
Secondary Structure
| Using primers VR/VF2 to PCR B0010 will result in excess bands.
See a full description of the problem here. |
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Measurement
- [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured]
>Internal Priming Screening Characterization of BBa_B0010: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
Examining efficiency of ribosomal RNA operon B (rrnB) T1 terminator from E. coli.: NYU Abu Dhabi

Transcription of DNA is made up of the following steps: RNA polymerase binding to promoter and activation, initiation of RNA transcript, elongation of RNA transcript, and termination of transcription. Termination is the last step of transcription where the RNA polymerase releases the RNA transcript. Releasing the RNA transcript is not reversible and further transcription requires reinitiation at a promoter region to form a new RNA transcript (Uptain et. al, 1997). In prokaryotes like E. coli, the termination sites serve as targets for gene expression regulation as they can not only occur at the end of genes, but also near promoter regions or between genes in the operon (Nojima et. al., 2005). The T1 terminator region on the rrnB gene of E. coli is one of two terminator regions, the other being T2. The two termination regions have "factor-independent terminator-like sequences" with "two additional inverted repeats (IR1 and IR2) and a pair of direct repeats" (Orosz et. al., 1991). The T1 and T2 terminating regions are often used as an efficient terminator of transcription in many cloning vectors (Orosz et. al., 1991).
Efficiency is enhanced by the E. coli nusA protein, which gives effectiveness of inhibition in vitro comparable to those in vivo. These transcripts that are terminated when nusA protein is present are released from the RNA polymerase complex, suggesting that there is a complete termination reaction. The protein's termination factor activity is not dependent on the presence ofthe rho protein. The nusA protein serves as an antitermination factor, RNA polymerase subunit, and true termination factor at some terminator sites. In general, termination at T1 in vitro is quiteefficient with an 80% effectiveness rate without any additional factors. In vivo and in E. coliextracts, the T1 terminator has shown to be nearly 100% efficient. In an isolated and purified system with only nusA protein present, termination at this high level of efficiency is also achieved, suggesting that in vivo, the entity responsible for the highly efficient termination is dueto the nusA protein.
Literature used:
- Nojima, T., A. C. Lin, T. Fujii and I. Endo (2005). "Determination of the Termination Efficiencyof the Transcription Terminator Using Different Fluorescent Profiles in Green Fluorescent Protein Mutants." Analytical Sciences 21(12): 1479-1481.Nojima, T.; Lin, A. C.; Fujii, T.; Endo,I., Determination of the Termination Efficiency of the Transcription Terminator Using Different Fluorescent Profiles in Green Fluorescent Protein Mutants. Analytical Sciences 2005, 21 (12), 1479-1481.
- Orosz, A., I. BOROS and P. VENETIANER (1991). "Analysis of the complex transcription termination region of the Escherichia coli rrn B gene." European journal of biochemistry 201(3): 653-659.
- Schmidt, M. C. and M. J. Chamberlin (1987). "nusA Protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites." Journal of Molecular Biology 195(4): 809-818.
- Uptain, S. M., & Chamberlin, M. J. (1997). Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proceedings of the National Academy of Sciences of the United States of America, 94(25), 13548–13553.
- Santangelo, T. J. and I. Artsimovitch (2011). "Termination and antitermination: RNA polymerase runs a stop sign." Nature reviews. Microbiology 9(5): 319-329.
This contribution was added by the 2020 NYU Abu Dhabi team.