Difference between revisions of "Part:BBa K3457014"
| Line 3: | Line 3: | ||
<partinfo>BBa_K3457014 short</partinfo> | <partinfo>BBa_K3457014 short</partinfo> | ||
| − | + | <p> This biological is the CDS sequence of secreted abundant heat soluble protein 33020, also called SAHS 33020. It is a kind of Tardigrade intrinsically Disordered Protein (TDP). It is a heat soluble protein found from Tardigrade <i>Hypsibius dujardini</i> in 2017 <sup>[1]</sup>. Tardigrade, also called water bear, is a kind of tenacious organism. It can survive extreme environment, such as desiccation, freeze and vacuum. The super capacity of Tardigrade partially owes to TDPs. Here we found that expressing this protein may help bacteria survive the freeze-drying process and then the resultant dry bacteria powder can be stored for a long time at room temperature. </p> | |
| + | |||
| + | ===Contribution=== | ||
| + | <h2><b>Group: QHFZ-China iGEM 2020</b></h2> | ||
| + | <h3><b>Author: Yixian Yang</b></h3> | ||
| + | <h3><b>Design</b></h3> | ||
| + | [[File:T--QHFZ-China--33020 basic design.png|400px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct of BBa_K3457012.]] | ||
| + | |||
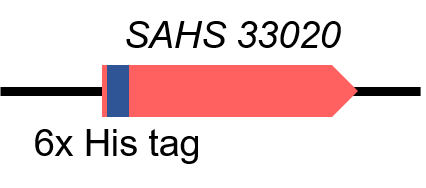
| + | <p style="clear:left;"> This part is the CDS of SAHS 33020. We obtained the sequence from the research paper in 2017 <sup>[1]</sup>. Then with the help of a company, we optimized the sequence, so that the part is suitable to work in <i>E. coli</i> and there is no biobrick restriction enzyme cutting site in it. Moreover, we added an 6x His tag at the N-terminal of SAHS 33020, so it can be easily detected by Western Blot and be purified by Ni-chelating affinity chromatography (Fig.1). This year, the part is used in the composite part [https://parts.igem.org/Part:BBa_K3457036 BBa_K3457036], [https://parts.igem.org/Part:BBa_K3457039 BBa_K3457039]</p> | ||
| + | |||
| + | <p style="clear:left;"> The part is similar to [https://parts.igem.org/Part:BBa_K2306003 BBa_K2306003], which is designed by iGEM17_TUDelft. However, no data were shown in the page and their wiki.</p> | ||
| + | |||
| + | <h3><b>Documentation:</b></h3> | ||
| + | <h4><b>Introduction: </b></h4> | ||
| + | <p style="clear:left;"> This year, we tried to introduce a new biopreservation method. We used freeze-drying to make the engineered into dry powder. Then the powder can be stored at room temperature for a long time. This method can make the storage of bacteria get rid of ultra-low temperature freezer, so that it will promote the practical application of engineered bacteria out of laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freeze, dry and vacuum, are lethal to bacteria. We use TDPs, including SAHS 33020, to help bacteria survive the situation. </p> | ||
| + | |||
| + | <h4><b>Protocol: </b></h4> | ||
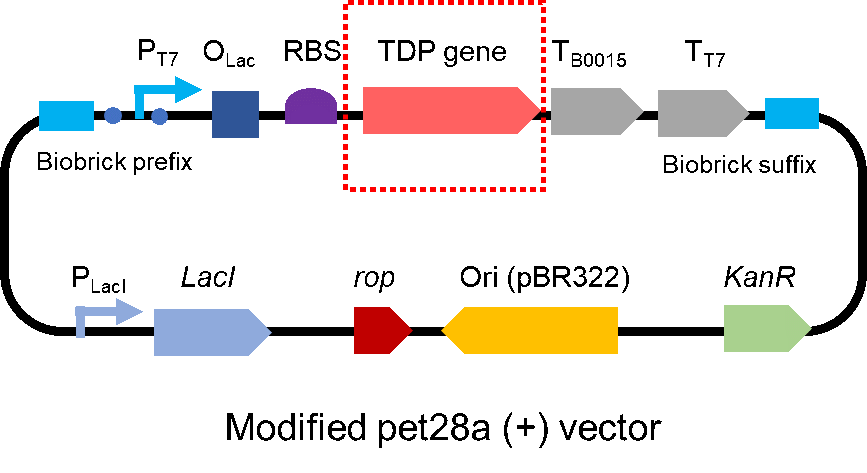
| + | <p style="clear:left;"> To test the effect of SAHS 33020, we modified a frequently and widely used vector, pet28a+ and put this part into it (Fig. 2). Then we transformed the plasmid into <i>E. coli</i> BL21 strain. </p> | ||
| + | |||
| + | [[File:T--QHFZ-China--pet28am.png|400px|thumb|left|Figure 2. The Schematic cartoon of the vector.]] | ||
| + | |||
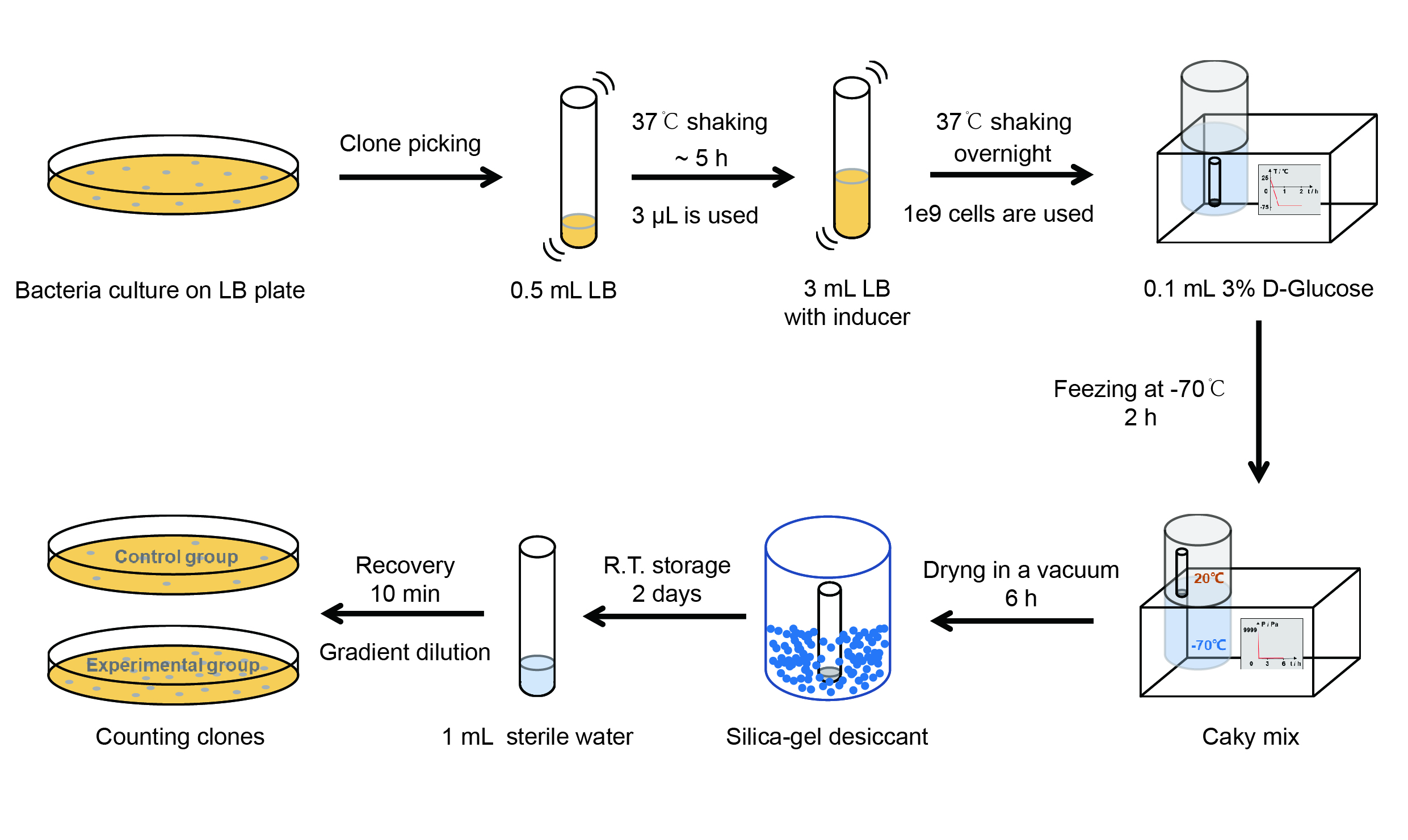
| + | <p style="clear:left;"> Then we used the following protocols to verify its function (Fig. 3): </p> | ||
| + | |||
| + | [[File:T--QHFZ-China--freeze-dry protocol.jpg|400px|thumb|left|Figure 3. Experiment protocol.]] | ||
| + | |||
| + | <p style="clear:left;"> | ||
| + | 【Day 1】Induction culture<br> | ||
| + | (1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid. <br> | ||
| + | (2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.<br> | ||
| + | 【Day 2】Freeze-dried<br> | ||
| + | (1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the experiment.<br> | ||
| + | (2) Use spectrophotometer to measure the OD<sub>600</sub> of the bacteria solution, OD<sub>600</sub> = 1 equals to 10<sup>9</sup> cells. If the OD<sub>600</sub> value is between 0.1 and 1, There is a linear relationship between OD<sub>600</sub> and bacterial density. Calculate the volume of bacterial solution for 10<sup>9</sup> cells by using the formula V = 100 / (OD<sub>600</sub> × Dilution ratio).<br> | ||
| + | (3) Take out a measured amount of 10<sup>9</sup> cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.<br> | ||
| + | (4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.<br> | ||
| + | (5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.<br> | ||
| + | (6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.<br> | ||
| + | (7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room temperature.<br> | ||
| + | 【Day 3】Room temperature storage<br> | ||
| + | 【Day 4】Detect the survival rate<br> | ||
| + | (1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.<br> | ||
| + | (2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on the LB plate.<br> | ||
| + | (3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several gradient dilutions.<br> | ||
| + | (4) Culture the bacteria overnight at 37℃.<br> | ||
| + | 【Day 5】Cell Count<br> | ||
| + | (1) Take out the LB plate and take photos to record experimental results.<br> | ||
| + | (2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the results between each group. | ||
| + | </p> | ||
| + | |||
| + | <h4><b>Results:</b></h4> | ||
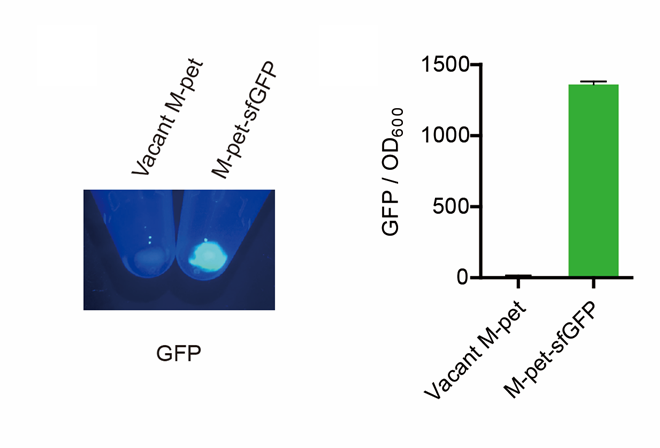
| + | <p style="clear:left;"> First, by a reporter, sfGFP, we confirmed that the plasmid can normally expressed exogenous proteins in <i>E. coli</i> BL21 strain (Fig. 4). Via SDS-PAGE, we observed the band of SAHS 33020, further verified the successfully expression (Fig.5). </p> | ||
| + | |||
| + | [[File:T--QHFZ-China--pet28am sfGFP.png|400px|thumb|left|Figure 4. Via a reporter, sfGFP, the expression function of the modified pet28a vector was verified.]] | ||
| + | [[File:T--QHFZ-China--TDP SDS.png|400px|thumb|left|Figure 5. SAHS 33020 was expressed as expected.]] | ||
| + | |||
| + | <p style="clear:left;"> Then, by freeze-drying and recover experiment, we tested the protective effect of SAHS 33020. However, in an independent repeat test, we found that compared with the control group (sfGFP), the bacteria expressing SAHS 33020 did not have a better survival rate (Fig. 6). </p> | ||
| + | |||
| + | [[File:T--QHFZ-China--TDP freeze-dry.png|400px|thumb|left|Figure 6. An independent repeat test of freeze-drying and recovery.]] | ||
| + | |||
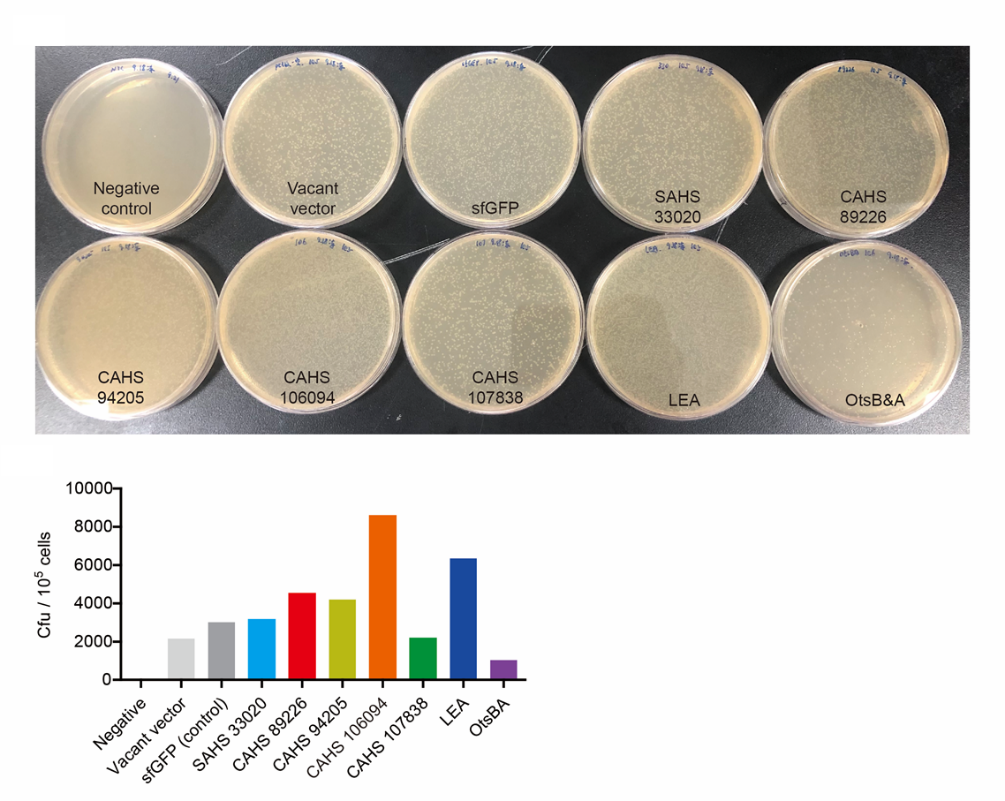
| + | <p style="clear:left;"> However, we thought that the result above was not credible. It was a little difficult for us to hold 10 samples for one time, so we thought maybe something was wrong. In some other independent repeat tests, bacteria expressing SAHS 33020 had a better survival rate than control group (Fig. 7). </p> | ||
| + | |||
| + | [[File:T--QHFZ-China--33020.png|400px|thumb|left|Figure 7. SAHS 33020 might protect bacteria during freeze-drying and subsequent dry storage]] | ||
| + | |||
| + | <h4 style="clear:left;"><b>Summary: </b></h4> | ||
| + | <p style="clear:left;"> SAHS 33020 may help the bacteria survive freeze-drying and subsequent dry storage. However, the effect were not steady. Further study should be taken to determine the function of SAHS 33020. You can also use the part to express and purify SAHS 33020 through Ni-chelating affinity chromatography. The purified SAHS 33020 can be used to protect protein products. </p> | ||
| + | |||
| + | <h4><b>Reference: </b></h4> | ||
| + | <p style="clear:left;">[1] Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., Rebecchi, L., Pielak, G.J., Koshland, D., and Goldstein, B. (2017). Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 65, 975-984 e975.</p><br> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 15:47, 25 October 2020
SAHS 33020 with a 6X His tag
This biological is the CDS sequence of secreted abundant heat soluble protein 33020, also called SAHS 33020. It is a kind of Tardigrade intrinsically Disordered Protein (TDP). It is a heat soluble protein found from Tardigrade Hypsibius dujardini in 2017 [1]. Tardigrade, also called water bear, is a kind of tenacious organism. It can survive extreme environment, such as desiccation, freeze and vacuum. The super capacity of Tardigrade partially owes to TDPs. Here we found that expressing this protein may help bacteria survive the freeze-drying process and then the resultant dry bacteria powder can be stored for a long time at room temperature.
Contribution
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
Design
This part is the CDS of SAHS 33020. We obtained the sequence from the research paper in 2017 [1]. Then with the help of a company, we optimized the sequence, so that the part is suitable to work in E. coli and there is no biobrick restriction enzyme cutting site in it. Moreover, we added an 6x His tag at the N-terminal of SAHS 33020, so it can be easily detected by Western Blot and be purified by Ni-chelating affinity chromatography (Fig.1). This year, the part is used in the composite part BBa_K3457036, BBa_K3457039
The part is similar to BBa_K2306003, which is designed by iGEM17_TUDelft. However, no data were shown in the page and their wiki.
Documentation:
Introduction:
This year, we tried to introduce a new biopreservation method. We used freeze-drying to make the engineered into dry powder. Then the powder can be stored at room temperature for a long time. This method can make the storage of bacteria get rid of ultra-low temperature freezer, so that it will promote the practical application of engineered bacteria out of laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freeze, dry and vacuum, are lethal to bacteria. We use TDPs, including SAHS 33020, to help bacteria survive the situation.
Protocol:
To test the effect of SAHS 33020, we modified a frequently and widely used vector, pet28a+ and put this part into it (Fig. 2). Then we transformed the plasmid into E. coli BL21 strain.
Then we used the following protocols to verify its function (Fig. 3):
【Day 1】Induction culture
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1 to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
【Day 2】Freeze-dried
(1) If fluorescence induced by the iPTG is detectable in the control group (GFP), continue conducting the experiment.
(2) Use spectrophotometer to measure the OD600 of the bacteria solution, OD600 = 1 equals to 109 cells. If the OD600 value is between 0.1 and 1, There is a linear relationship between OD600 and bacterial density. Calculate the volume of bacterial solution for 109 cells by using the formula V = 100 / (OD600 × Dilution ratio).
(3) Take out a measured amount of 109 cells and centrifuge it at 8000 rpm for 3 min. Then pour out the supernatant.
(4) Resuspend the bacteria in a 15 mL tube with pre-refrigerated 100 μL 3% glucose solution.
(5) Take off the cover of the tube and put the bacteria into the cold trap. Open the compressor of the lyophilization machine and freeze the shake tube for 2 h at -70℃.
(6) Put the caky bacteria solution into the drying chamber of the lyophilization machine. Open the vacuum pump to dry it in vacuum for 6h at 1 Pa vacuum degree.
(7) Turn off the vacuum pump, place it at seal box filled with silica-gel desiccant a for 2 days at room temperature.
【Day 3】Room temperature storage
【Day 4】Detect the survival rate
(1) Add 1 mL of sterile water to the tube, vortex for 15 s, placed it at room temperature for 10 min.
(2) Adjust the density of the bacteria solution by gradient dilution, then spread 100 μL of the bacteria solution on the LB plate.
(3) If the density above is not suitable, take 100μL of the solution and spread it on the LB plate after several gradient dilutions.
(4) Culture the bacteria overnight at 37℃.
【Day 5】Cell Count
(1) Take out the LB plate and take photos to record experimental results.
(2) Use the automatic cell counting function of Image J to count the colone number on the LB plate, then compare the results between each group.
Results:
First, by a reporter, sfGFP, we confirmed that the plasmid can normally expressed exogenous proteins in E. coli BL21 strain (Fig. 4). Via SDS-PAGE, we observed the band of SAHS 33020, further verified the successfully expression (Fig.5).
Then, by freeze-drying and recover experiment, we tested the protective effect of SAHS 33020. However, in an independent repeat test, we found that compared with the control group (sfGFP), the bacteria expressing SAHS 33020 did not have a better survival rate (Fig. 6).
However, we thought that the result above was not credible. It was a little difficult for us to hold 10 samples for one time, so we thought maybe something was wrong. In some other independent repeat tests, bacteria expressing SAHS 33020 had a better survival rate than control group (Fig. 7).
Summary:
SAHS 33020 may help the bacteria survive freeze-drying and subsequent dry storage. However, the effect were not steady. Further study should be taken to determine the function of SAHS 33020. You can also use the part to express and purify SAHS 33020 through Ni-chelating affinity chromatography. The purified SAHS 33020 can be used to protect protein products.
Reference:
[1] Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., Rebecchi, L., Pielak, G.J., Koshland, D., and Goldstein, B. (2017). Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol Cell 65, 975-984 e975.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]