Difference between revisions of "Part:BBa K538101"
CaptainCold (Talk | contribs) |
|||
| (4 intermediate revisions by one other user not shown) | |||
| Line 20: | Line 20: | ||
<p>As part of charecterization, the iGEM IISER Bhopal 2019 team performed growth curves at different temperatures in order to study the part which was also a part of our project. </p> | <p>As part of charecterization, the iGEM IISER Bhopal 2019 team performed growth curves at different temperatures in order to study the part which was also a part of our project. </p> | ||
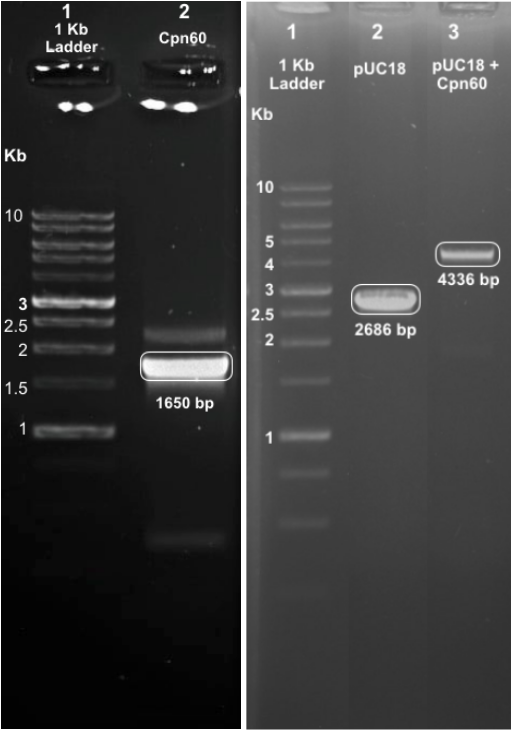
| − | <p>Cpn 60 was successfully digested from the parent backbone pSB1C3, which was confirmed on gel via size confirmation and sequencing exercises | + | <p>Cpn 60 was successfully digested from the parent backbone pSB1C3, which was confirmed on gel via size confirmation and sequencing exercises and inserted into pUC18</p> |
| + | https://static.igem.org/mediawiki/parts/e/e2/T--IISERB_Bhopal--Cpn60_lowppx.png | ||
https://2019.igem.org/wiki/images/d/d3/T--IISER_Bhopal--cpn60cloneresult.png | https://2019.igem.org/wiki/images/d/d3/T--IISER_Bhopal--cpn60cloneresult.png | ||
As this part had already been submitted by the 2011 Amsterdam team, we decided to further characterize it (as a part of the Bronze medal criteria) and check how its induction affected cell growth. The results of the growth analysis may be seen in the graphs provided below. The temperatures chosen were 37°C (as a model for optimal conditions) and 18°C (as a model for suboptimal conditions). While we ideally would have liked to achieve even lower temperatures (as our model clearly predicts that TF could survive near 0°C-like conditions), we were most unfortunately limited by the available infrastructural facilities.</p> | As this part had already been submitted by the 2011 Amsterdam team, we decided to further characterize it (as a part of the Bronze medal criteria) and check how its induction affected cell growth. The results of the growth analysis may be seen in the graphs provided below. The temperatures chosen were 37°C (as a model for optimal conditions) and 18°C (as a model for suboptimal conditions). While we ideally would have liked to achieve even lower temperatures (as our model clearly predicts that TF could survive near 0°C-like conditions), we were most unfortunately limited by the available infrastructural facilities.</p> | ||
| − | https:// | + | https://static.igem.org/mediawiki/parts/4/46/T--IISERB_Bhopal--_Cpn60_g1_lowppx.png https://static.igem.org/mediawiki/parts/6/61/T--IISERB_Bhopal--_Cpn60_g2_lowppx.png |
| − | |||
| + | For confirmation, we also ran a whole cell lysate SDS page with induced Cpn60 cells, this gave a distinct band at approxiamtely 60kDa (actual weight of the protein is 56,872 Da). The gel image is as seen below<br> | ||
| + | https://static.igem.org/mediawiki/parts/8/8f/T--IISERB_Bhopal--_SDS_lowppx.png | ||
===Sequence and Features=== | ===Sequence and Features=== | ||
<partinfo>K538101 SequenceAndFeatures</partinfo> | <partinfo>K538101 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | ==Functional Parameters: Austin_UTexas== | ||
| + | <html> | ||
| + | <body> | ||
| + | <partinfo>BBa_K538101 parameters</partinfo> | ||
| + | <h3><center>Burden Imposed by this Part:</center></h3> | ||
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 1.1 ± 4.7% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 19:04, 27 August 2020
RBS + Cpn60
This part was submitted by team [http://2011.igem.org/Team:Amsterdam Amsterdam 2011] as part of their project: icE. coli. It was used as a construction intermediate, to create the following composite parts:
For more information about this brick's biology, refer to the page of its coding region: BBa_K538001
Results
As part of charecterization, the iGEM IISER Bhopal 2019 team performed growth curves at different temperatures in order to study the part which was also a part of our project.
Cpn 60 was successfully digested from the parent backbone pSB1C3, which was confirmed on gel via size confirmation and sequencing exercises and inserted into pUC18

As this part had already been submitted by the 2011 Amsterdam team, we decided to further characterize it (as a part of the Bronze medal criteria) and check how its induction affected cell growth. The results of the growth analysis may be seen in the graphs provided below. The temperatures chosen were 37°C (as a model for optimal conditions) and 18°C (as a model for suboptimal conditions). While we ideally would have liked to achieve even lower temperatures (as our model clearly predicts that TF could survive near 0°C-like conditions), we were most unfortunately limited by the available infrastructural facilities.</p>


For confirmation, we also ran a whole cell lysate SDS page with induced Cpn60 cells, this gave a distinct band at approxiamtely 60kDa (actual weight of the protein is 56,872 Da). The gel image is as seen below

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 888
Illegal BamHI site found at 954 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.
