Difference between revisions of "Part:BBa K2916048"
Konstantinos (Talk | contribs) |
Konstantinos (Talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2916048 short</partinfo> | <partinfo>BBa_K2916048 short</partinfo> | ||
| − | This part is used for expression of | + | This part is used for expression of Inorganic pyrophosphatasePiase needed for the OnePot PURE cell-free system. |
<!-- Add more about the biology of this part here--> | <!-- Add more about the biology of this part here--> | ||
| Line 9: | Line 9: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | Inorganic pyrophosphatase (PPiase) is responsible for regulating the secondary structure of the expressed proteins. | |
| + | |||
| + | In our project we used PPiase as a part of the protein solution needed for <html><a style="padding: 0px; margin: 0px;" href="https://2019.igem.org/Team:EPFL/OnePot_Pure"> OnePot PURE cell-free system </a></html> produced with the method of gravity flow affinity chromatography, as described in the <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/protein-purification-for-onepot-pure-cell-free-sys-8auhsew"> protocol </a></html> we designed. | ||
| + | |||
| + | ===Characterization=== | ||
| + | |||
| + | =='''Expression and purification of PPiase'''== | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | PPiase is one of the proteins we used for the OnePot PURE cell-free system. We expressed it in BL21(DE3) E.coli strain using a pET21a vector. The expression system has a T7 promoter, a lac operator, RBS and a T7 Terminator, enabling us to regulate the expression with IPTG.<br/><br/> | ||
| + | '''Methods''' | ||
| + | <br/> | ||
| + | |||
| + | PPiase was purified using our <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/protein-purification-for-onepot-pure-cell-free-sys-8auhsew"> protocol </a></html>. To test if the protein was actually expressed, we performed a SDS-PAGE that is presented below. On the left side we can see the results included in the initial OnePot PURE paper (<i>Lavickova et al, 2019</i>) while on the right (batch1_a,b and batch2_a,b) are the solutions we produced ourselves. (The procedure we followed and the conditions of the experiment can be found <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/sps-page-protein-electrophoresis-775hrq6"> here</a></html>). | ||
| + | <br/> | ||
| + | |||
| + | <html> | ||
| + | <figure style="text-align:center;"> | ||
| + | <img style="max-width:700px;" src="https://2019.igem.org/wiki/images/b/bb/T--EPFL--ProteinMolec.png" alt="control"> | ||
| + | <figcaption><b>Figure 1:</b> SDS-PAGE of OnePot PURE protein solution.</figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| + | '''Conclusion''' | ||
| + | <br/> PPiase has a molecular weight of around 32kDa, but even though we cannot be absolutely sure if the band shown is only due to it, we may assume that it is expressed. To verify the existence and functionality of this protein we need to proceed with more experiments that would be mainly focused on the efficiency of the system. | ||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | =='''OnePot PURE functionality test'''== | ||
| + | |||
| + | |||
| + | <br/> | ||
| + | To make sure that we have all the proteins in our OnePot PURE protein solution, and that they all function properly we need check if proteins can be expressed in our OnePot PURE cell-free system. <br/> | ||
| + | |||
| + | '''Methods'''<br/> | ||
| + | |||
| + | We expressed <html><a style="padding: 0px; margin: 0px;" href="https://parts.igem.org/Part:BBa_I746909"> superfolding GFP</a></html> following the <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/protein-expression-in-onepot-pure-cell-free-system-8avhse6"> protocol</a></html> we designed in 10μl reactions, and measured the fluorescence on a plate reader at excitation wavelength of 535nm. We tested the expression using different concentrations of the sf GFP DNA template and also compared it with the fluorescence produced in PURExpress from NEB. | ||
| + | <br/> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <figure style="text-align:center;"> | ||
| + | <img style="max-width:700px;" src="https://2019.igem.org/wiki/images/b/b2/T--EPFL--resultsOnePot10.png" alt="control"> | ||
| + | <figcaption><b>Figure 2:</b> sf GFP expression using 10nM DNA template.</figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | <html> | ||
| + | <figure style="text-align:center;"> | ||
| + | <img style="max-width:700px;" src="https://2019.igem.org/wiki/images/7/75/T--EPFL--resultsOnePot5.png" alt="control"> | ||
| + | <figcaption><b>Figure 3:</b> sf GFP expression using 5nM DNA template.</figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | <html> | ||
| + | <figure style="text-align:center;"> | ||
| + | <img style="max-width:700px;" src="https://2019.igem.org/wiki/images/e/e7/T--EPFL--resultsOnePot25.png" alt="control"> | ||
| + | <figcaption><b>Figure 4:</b> sf GFP expression using 2.5nM DNA template.</figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | <html> | ||
| + | <figure style="text-align:center;"> | ||
| + | <img style="max-width:700px;" src="https://2019.igem.org/wiki/images/7/70/T--EPFL--resultsOnePot.png" alt="control"> | ||
| + | <figcaption><b>Figure 5:</b> Comparison between OnePot PURE and PURExpress at saturation.</figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| + | '''Conclusion''' | ||
| + | <br/> The expression was successful so we can confirm that PPiase exists in our protein solution and is also functioning properly. | ||
| + | <br/> | ||
| + | |||
| + | |||
Latest revision as of 03:34, 22 October 2019
Expression of PPiase in E.coli
This part is used for expression of Inorganic pyrophosphatasePiase needed for the OnePot PURE cell-free system.
Usage and Biology
Inorganic pyrophosphatase (PPiase) is responsible for regulating the secondary structure of the expressed proteins.
In our project we used PPiase as a part of the protein solution needed for OnePot PURE cell-free system produced with the method of gravity flow affinity chromatography, as described in the protocol we designed.
Characterization
Expression and purification of PPiase
PPiase is one of the proteins we used for the OnePot PURE cell-free system. We expressed it in BL21(DE3) E.coli strain using a pET21a vector. The expression system has a T7 promoter, a lac operator, RBS and a T7 Terminator, enabling us to regulate the expression with IPTG.
Methods
PPiase was purified using our protocol . To test if the protein was actually expressed, we performed a SDS-PAGE that is presented below. On the left side we can see the results included in the initial OnePot PURE paper (Lavickova et al, 2019) while on the right (batch1_a,b and batch2_a,b) are the solutions we produced ourselves. (The procedure we followed and the conditions of the experiment can be found here).
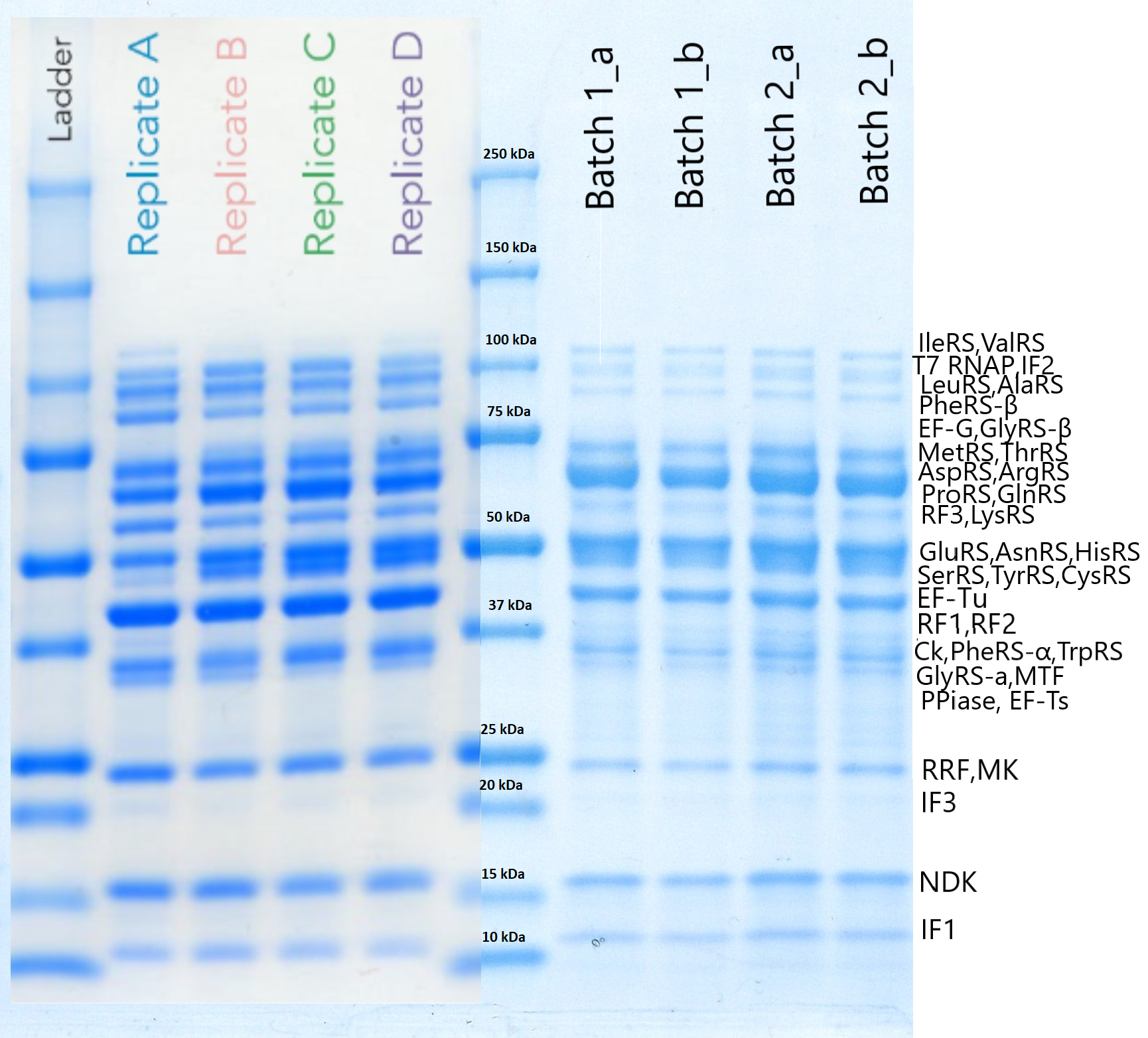
Conclusion
PPiase has a molecular weight of around 32kDa, but even though we cannot be absolutely sure if the band shown is only due to it, we may assume that it is expressed. To verify the existence and functionality of this protein we need to proceed with more experiments that would be mainly focused on the efficiency of the system.
OnePot PURE functionality test
To make sure that we have all the proteins in our OnePot PURE protein solution, and that they all function properly we need check if proteins can be expressed in our OnePot PURE cell-free system.
Methods
We expressed superfolding GFP following the protocol we designed in 10μl reactions, and measured the fluorescence on a plate reader at excitation wavelength of 535nm. We tested the expression using different concentrations of the sf GFP DNA template and also compared it with the fluorescence produced in PURExpress from NEB.
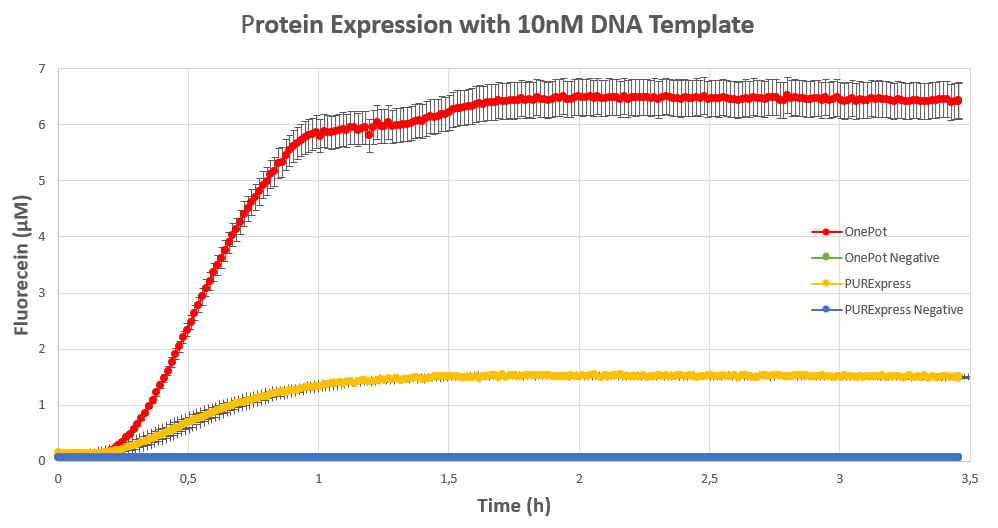
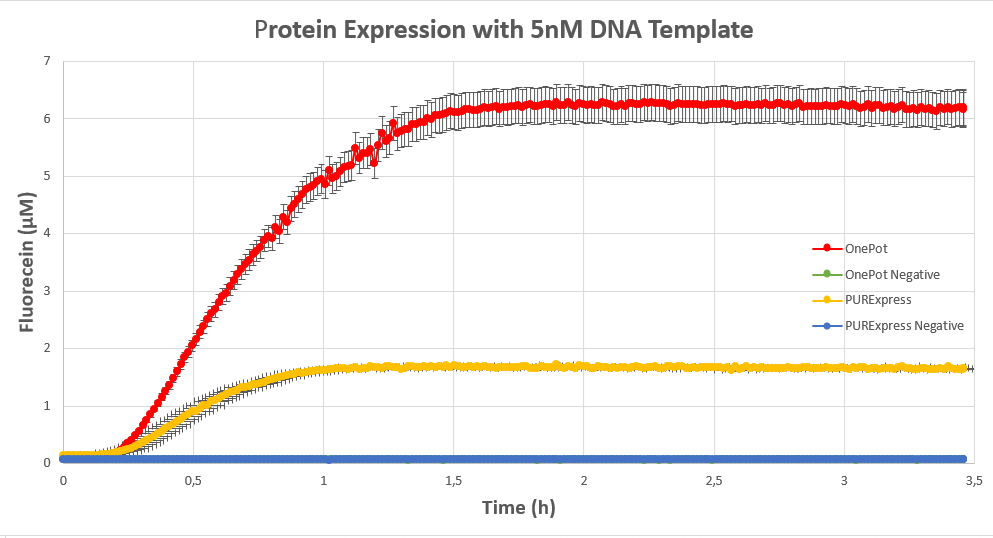
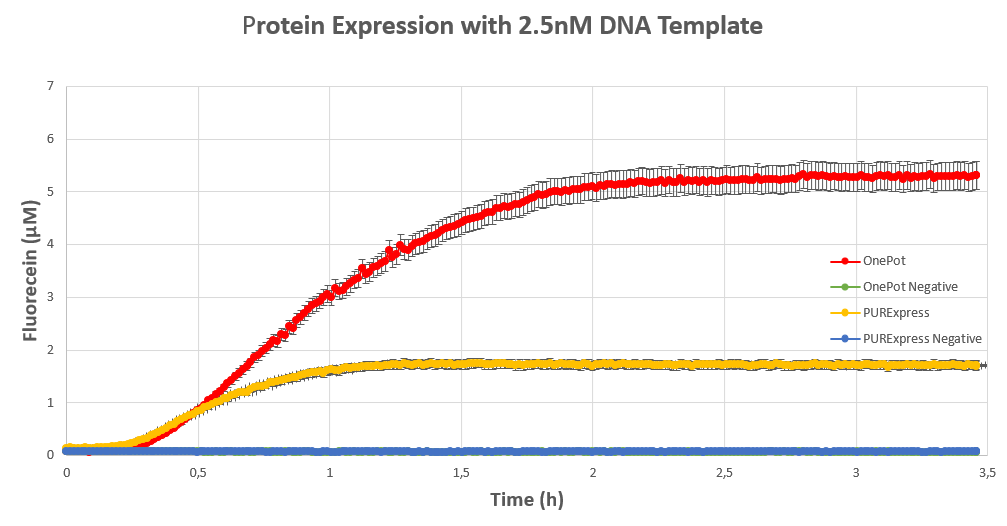
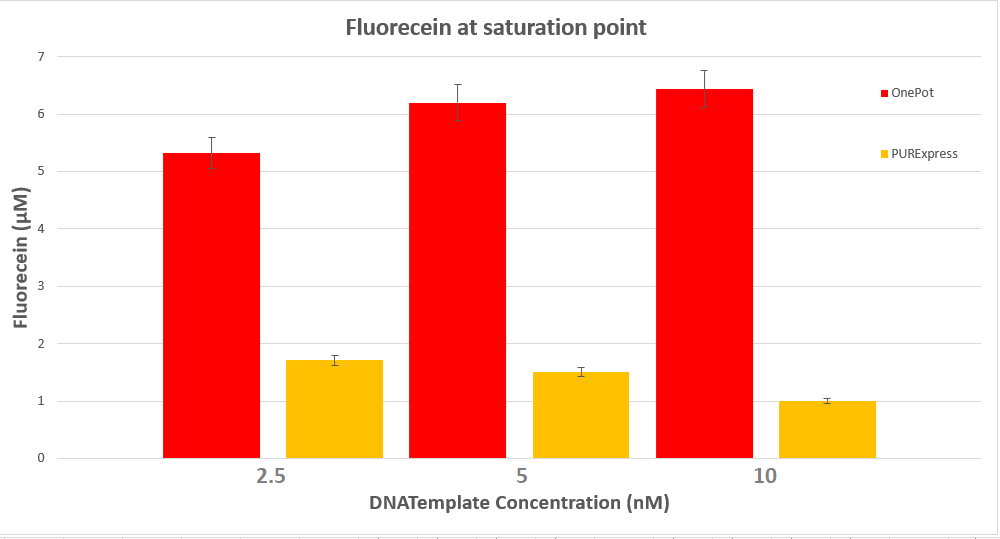
Conclusion
The expression was successful so we can confirm that PPiase exists in our protein solution and is also functioning properly.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1007
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 956
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 545
