Difference between revisions of "Part:BBa K3254014"
Zongyeqing (Talk | contribs) |
(→Thermodynamic Characterization) |
||
| Line 8: | Line 8: | ||
=Thermodynamic Characterization= | =Thermodynamic Characterization= | ||
*You can find the same contents below on the page of [[Part:BBa_K2572025|BBa_K2572025]]. | *You can find the same contents below on the page of [[Part:BBa_K2572025|BBa_K2572025]]. | ||
| − | *This part can be seen as an improved version of the original Ptac promoter ([[Part:BBa_K864400|BBa_K864400]]). The main improvement point | + | *This part can be seen as an improved version of the original Ptac promoter ([[Part:BBa_K864400|BBa_K864400]]). The main improvement point was this version could be applied a symmetrical lacO site to get a more tightly regulation. |
| − | *The page of a twin part was | + | *The page of a twin part was build for a full annotation ([[Part:BBa_K3254014|BBa_K3254014]]). |
*We used this part to construct an IPTG inducible transcriptional device by combining with a lacI expression cassette([[Part:BBa_K3254022|BBa_K3254022]]) on a P15A plamisd. An insulated sfgfp reporter translational unit ([[Part:BBa_K3254024|BBa_K3254024]]) was applied for quantitative assay the dynamic response curve. The full structure of this operon was described on the page of [[Part:BBa_K3254025|BBa_K3254025]]. | *We used this part to construct an IPTG inducible transcriptional device by combining with a lacI expression cassette([[Part:BBa_K3254022|BBa_K3254022]]) on a P15A plamisd. An insulated sfgfp reporter translational unit ([[Part:BBa_K3254024|BBa_K3254024]]) was applied for quantitative assay the dynamic response curve. The full structure of this operon was described on the page of [[Part:BBa_K3254025|BBa_K3254025]]. | ||
*Furthermore, we improved this part by mutant the -10 region of this promoter and successfully achieved a series of IPTG inducible promoters([[Part:BBa_K3254015|Ptac-M1]], [[Part:BBa_K3254016|Ptac-M2]], [[Part:BBa_K3254017|Ptac-M3]]) with different dynamic ranges and lower non-induced activities. | *Furthermore, we improved this part by mutant the -10 region of this promoter and successfully achieved a series of IPTG inducible promoters([[Part:BBa_K3254015|Ptac-M1]], [[Part:BBa_K3254016|Ptac-M2]], [[Part:BBa_K3254017|Ptac-M3]]) with different dynamic ranges and lower non-induced activities. | ||
| Line 16: | Line 16: | ||
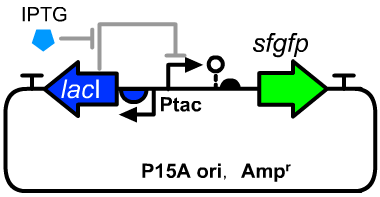
[[File:T--GENAS_China--Ptac-sfGFP_plasmid.PNG|200px|thumb|left|The structure of the experimental plasmid]] | [[File:T--GENAS_China--Ptac-sfGFP_plasmid.PNG|200px|thumb|left|The structure of the experimental plasmid]] | ||
*The sequence detail was described on the page of [[Part:BBa_K3254025|BBa_K3254025]]. | *The sequence detail was described on the page of [[Part:BBa_K3254025|BBa_K3254025]]. | ||
| − | *The host cell | + | *The host cell was E.coli DH5α. |
==Experimental Setup== | ==Experimental Setup== | ||
| − | *All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using Corning flat-bottom 96-well plates sealed with sealing film. For | + | *All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using Corning flat-bottom 96-well plates sealed with sealing film. For characterizing the circuit response functions, a previously developed quantitative method that measures gene expression at steady state was used(Zhang, Chen et al. 2016). Briefly, bacteria harboring the parts/circuits of interest were first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth, after which the cell cultures were diluted 196-fold with M9 medium. After 3 h of growth, the cultures were further diluted 700-fold with M9 medium containing gradient concentrations of IPTG, and incubated for another 6 h. Finally, 20-μL samples of each culture were transferred to a new plate containing 180 μL per well of PBS supplemented with 2 mg/mL kanamycin to terminate protein expression. The fluorescence distribution of each sample was assayed using a flow cytometer with appropriate voltage settings; each distribution contained >20,000 events. Each sample was experimentally assayed at least three times. The arithmetical mean of each sample was determined using FlowJo software. |
*M9 medium (supplemented): 6.8 g/L Na<sub>2</sub>HPO<sub>4</sub>, 3 g/L KH<sub>2</sub>PO<sub>4</sub>, 0.5 g/L NaCl, 1 g/L NH<sub>4</sub>Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO<sub>4</sub>, and 100 μM CaCl<sub>2</sub>. | *M9 medium (supplemented): 6.8 g/L Na<sub>2</sub>HPO<sub>4</sub>, 3 g/L KH<sub>2</sub>PO<sub>4</sub>, 0.5 g/L NaCl, 1 g/L NH<sub>4</sub>Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO<sub>4</sub>, and 100 μM CaCl<sub>2</sub>. | ||
Revision as of 11:39, 21 October 2019
Optimized Ptac promoter
This part is a twin of BBa_K2572025, we built this page just for more detailed sequence annotations. This promoter was optimized from Ptac promoter(BBa_K864400) through using a symmetrical lacO site. After combined with the lacI constitutive expression cassette(BBa_K3254022), it can be regulated by the IPTG inducer. A mutant library with different dynamic range had been built(see BBa_K3254015,BBa_K3254016 and BBa_K3254017).
Thermodynamic Characterization
- You can find the same contents below on the page of BBa_K2572025.
- This part can be seen as an improved version of the original Ptac promoter (BBa_K864400). The main improvement point was this version could be applied a symmetrical lacO site to get a more tightly regulation.
- The page of a twin part was build for a full annotation (BBa_K3254014).
- We used this part to construct an IPTG inducible transcriptional device by combining with a lacI expression cassette(BBa_K3254022) on a P15A plamisd. An insulated sfgfp reporter translational unit (BBa_K3254024) was applied for quantitative assay the dynamic response curve. The full structure of this operon was described on the page of BBa_K3254025.
- Furthermore, we improved this part by mutant the -10 region of this promoter and successfully achieved a series of IPTG inducible promoters(Ptac-M1, Ptac-M2, Ptac-M3) with different dynamic ranges and lower non-induced activities.
Genetic Design
- The sequence detail was described on the page of BBa_K3254025.
- The host cell was E.coli DH5α.
Experimental Setup
- All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using Corning flat-bottom 96-well plates sealed with sealing film. For characterizing the circuit response functions, a previously developed quantitative method that measures gene expression at steady state was used(Zhang, Chen et al. 2016). Briefly, bacteria harboring the parts/circuits of interest were first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth, after which the cell cultures were diluted 196-fold with M9 medium. After 3 h of growth, the cultures were further diluted 700-fold with M9 medium containing gradient concentrations of IPTG, and incubated for another 6 h. Finally, 20-μL samples of each culture were transferred to a new plate containing 180 μL per well of PBS supplemented with 2 mg/mL kanamycin to terminate protein expression. The fluorescence distribution of each sample was assayed using a flow cytometer with appropriate voltage settings; each distribution contained >20,000 events. Each sample was experimentally assayed at least three times. The arithmetical mean of each sample was determined using FlowJo software.
- M9 medium (supplemented): 6.8 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO4, and 100 μM CaCl2.
Results
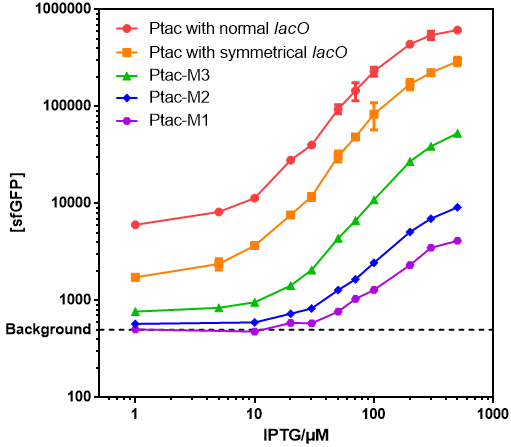
- Those promoters have gentle response curves without saltus.
- Background: Cells without any FP genes.
- Ptac with normal lacO: BBa_K864400
- Ptac with symmetrical lacO: BBa_K2572025
- Ptac-M1: BBa_K3254015
- Ptac-M2: BBa_K3254016
- Ptac-M3: BBa_K3254017
References
- Zhang HM, et al. Measurements of Gene Expression at Steady State Improve the Predictability of Part Assembly. ACS Synthetic Biology 5, 269-273 (2016).
- Zong Y, et al. Insulated transcriptional elements enable precise design of genetic circuits. Nature communications 8, 52 (2017).
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]