Difference between revisions of "Part:BBa K3147006"
(→I. Part BBa_K3147006 function) |
|||
| (12 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <partinfo>BBa_K3147006 short</partinfo> | + | <div align="center"><partinfo>BBa_K3147006 short</partinfo></div> |
| + | |||
| + | |||
| + | |||
| + | ===I. Part BBa_K3147006 function=== | ||
| + | |||
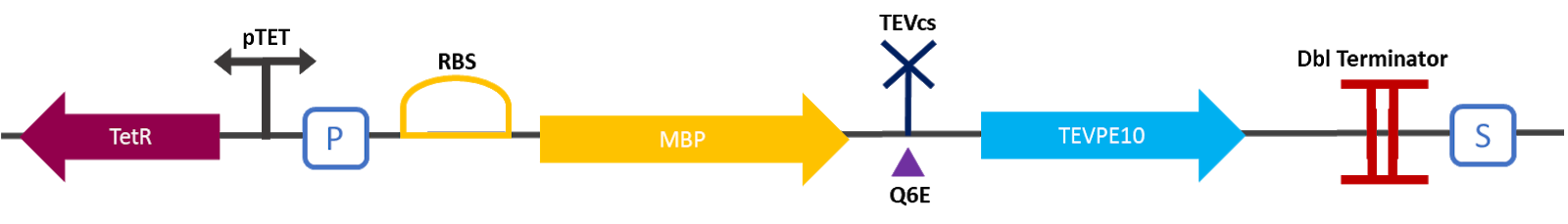
| + | The 2019 Montpellier iGEM team made this construction in order to be able to compare the basal activity of the TEV protease [[Part:BBa_ K1319004]] with the mutant TEV protease TEVPE10 [3]. A modified TEV cut-off site was added between the MBP and the protease. MBP increases the solubility of the fusion protein [1], preventing the aggregation of the protein of interest. This stabilizes the expression and the sequence of the produced MBP does not have a signal peptide, which allows the protein to remain in the cytosol. The TEV cutting site allows self-cleavage of MBP from TEV once the protein is produced [2]. | ||
| + | |||
| + | <div align="center">[[File:designK3147006.png|650px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 1</b>: Construct Design: MBP-TEVcs-TEVPE10 with modified TEV cutting site.</div> | ||
| + | |||
| + | ===II. Proof of function=== | ||
| + | |||
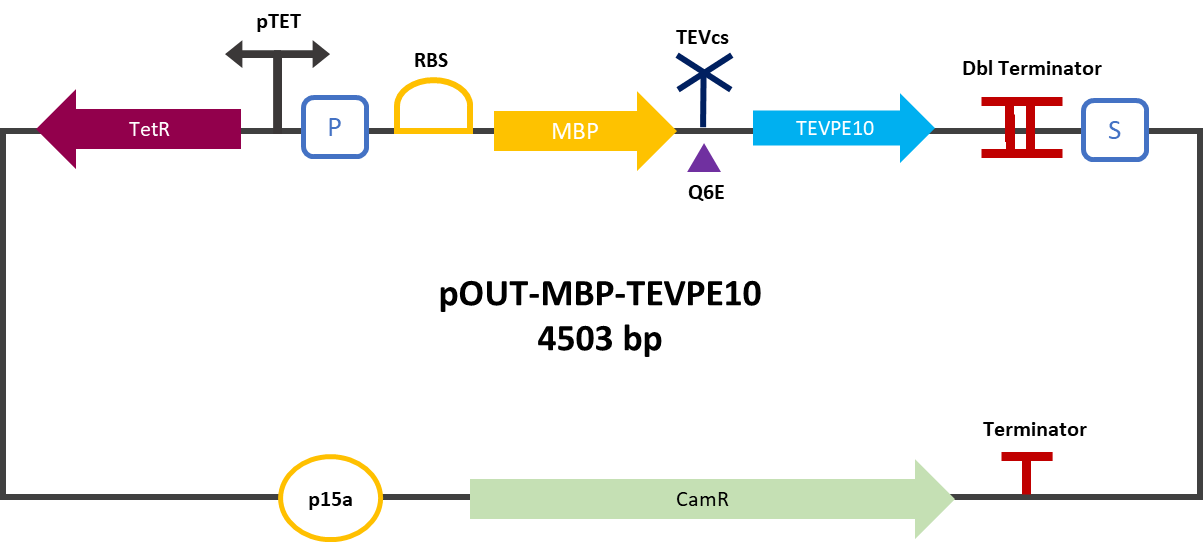
| + | The construction was cloned by Gibson Assembly in a pOUT18 backbone under the control of a Tet-on promoter in order to control its expression. The TEV cutting site used is the mutated site: ENLYFE/G this mutation allows theoretically the mutant TEV to better cleave this sequence according to this source [3]. | ||
| + | |||
| + | <div align="center">[[File:plasmideK3147006.png|450px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 2</b> : MBP-TEVPE10 reporter gene in its backbone pOUT18.</div> | ||
| + | |||
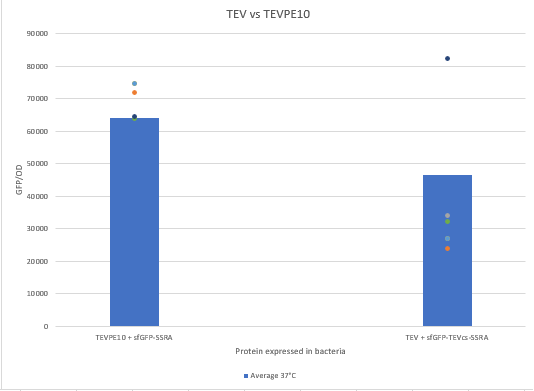
| + | The experimental approach that has been used to test the protease activity is to compare the fluorescence restoration rate of sfGFP compared to MBP-TEV and MBP-TEVPE10. In this experiment, basal controls of maximum and minimum fluorescence of the reporter genes were used. Fluorescence data are obtained from plate reader at 37°C with <i>E.coli</i> NEB10B. The protease is expressed by inducing the Tet promoter with 50 ng/mL of aTc (anhydrotetracycline). | ||
| + | |||
| + | <div align="center">[[File:resultK3147006.png|500px]]</div> | ||
| + | |||
| + | <div align="center"><b>Figure 2</b>: Comparison of the activity of TEVPE10 to the wild-type TEV by measuring the fluorescence of a reporter fused to a cleavable proteolysis tag</div> | ||
| + | |||
| + | We can see that the TEVPE10 is more efficient than the wild-type TEV. We believe this is because our wild-type TEV is not the same one in the article where we found this mutant. | ||
| + | |||
| + | ==Reference== | ||
| + | |||
| + | [1]Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589. | ||
| + | |||
| + | [2]Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41. | ||
| + | |||
| + | [3] Yi, L. et al. 2013. « Engineering of TEV Protease Variants by Yeast ER Sequestration Screening (YESS) of Combinatorial Libraries ». Proceedings of the National Academy of Sciences 110(18): 7229‑34. | ||
| + | |||
| − | |||
| − | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 16:33, 20 October 2019
I. Part BBa_K3147006 function
The 2019 Montpellier iGEM team made this construction in order to be able to compare the basal activity of the TEV protease Part:BBa_ K1319004 with the mutant TEV protease TEVPE10 [3]. A modified TEV cut-off site was added between the MBP and the protease. MBP increases the solubility of the fusion protein [1], preventing the aggregation of the protein of interest. This stabilizes the expression and the sequence of the produced MBP does not have a signal peptide, which allows the protein to remain in the cytosol. The TEV cutting site allows self-cleavage of MBP from TEV once the protein is produced [2].
II. Proof of function
The construction was cloned by Gibson Assembly in a pOUT18 backbone under the control of a Tet-on promoter in order to control its expression. The TEV cutting site used is the mutated site: ENLYFE/G this mutation allows theoretically the mutant TEV to better cleave this sequence according to this source [3].
The experimental approach that has been used to test the protease activity is to compare the fluorescence restoration rate of sfGFP compared to MBP-TEV and MBP-TEVPE10. In this experiment, basal controls of maximum and minimum fluorescence of the reporter genes were used. Fluorescence data are obtained from plate reader at 37°C with E.coli NEB10B. The protease is expressed by inducing the Tet promoter with 50 ng/mL of aTc (anhydrotetracycline).
We can see that the TEVPE10 is more efficient than the wild-type TEV. We believe this is because our wild-type TEV is not the same one in the article where we found this mutant.
Reference
[1]Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589.
[2]Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41.
[3] Yi, L. et al. 2013. « Engineering of TEV Protease Variants by Yeast ER Sequestration Screening (YESS) of Combinatorial Libraries ». Proceedings of the National Academy of Sciences 110(18): 7229‑34.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 77
Illegal SapI.rc site found at 1868