Difference between revisions of "Part:BBa K3221205"
(→iGEM19_SCU-China:) |
(→iGEM19_SCU-China:) |
||
| Line 19: | Line 19: | ||
<!-- --> | <!-- --> | ||
=='''iGEM19_SCU-China:'''== | =='''iGEM19_SCU-China:'''== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p style="font-size:18px !important;">The yamilCP was improved from amilCP (Part: [https://parts.igem.org/Part:BBa_K592009 BBa_K592009]), which is a blue chromoprotein gene from <i>Acropora millepora</i> submitted by Team Uppsala-Sweden 2011.</p> | <p style="font-size:18px !important;">The yamilCP was improved from amilCP (Part: [https://parts.igem.org/Part:BBa_K592009 BBa_K592009]), which is a blue chromoprotein gene from <i>Acropora millepora</i> submitted by Team Uppsala-Sweden 2011.</p> | ||
| − | <p>We expected amilCP to be expressed efficiently in <i>S. cerevisiae</i>, so we chose amilCP to be improved. We processed codon optimization on original amilCP sequence and added a consensus sequence (AAAAAA) for <i>S. cerevisiae</i> expression. And the new sequence was named yamilCP.</p> | + | <p style="font-size:18px !important;">We expected amilCP to be expressed efficiently in <i>S. cerevisiae</i>, so we chose amilCP to be improved. We processed codon optimization on original amilCP sequence and added a consensus sequence (AAAAAA) for <i>S. cerevisiae</i> expression. And the new sequence was named yamilCP.</p> |
<p>[[File:T--SCU-China--scu-improvement-Figure1.png|450px||left|]] | <p>[[File:T--SCU-China--scu-improvement-Figure1.png|450px||left|]] | ||
| Line 46: | Line 41: | ||
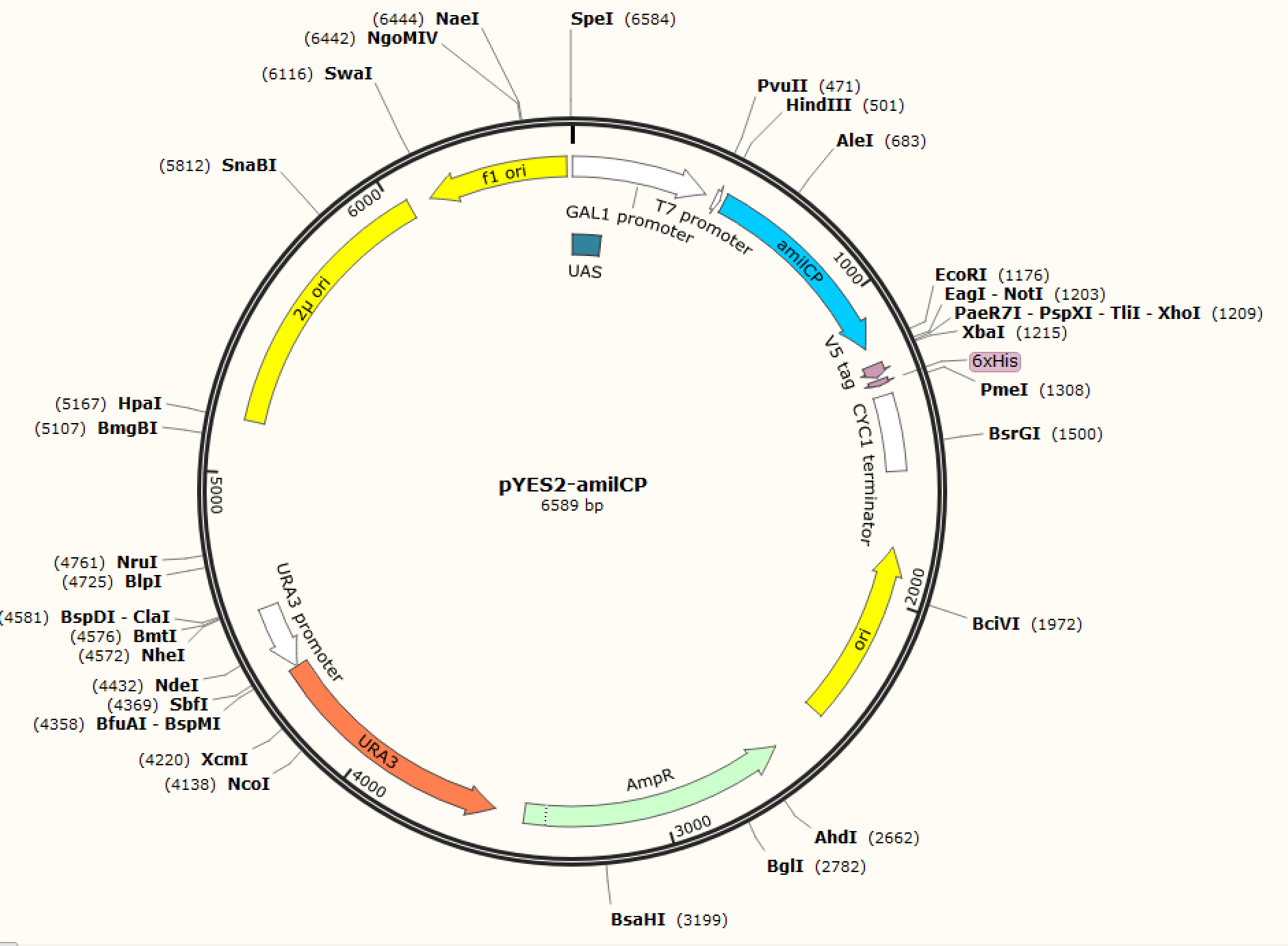
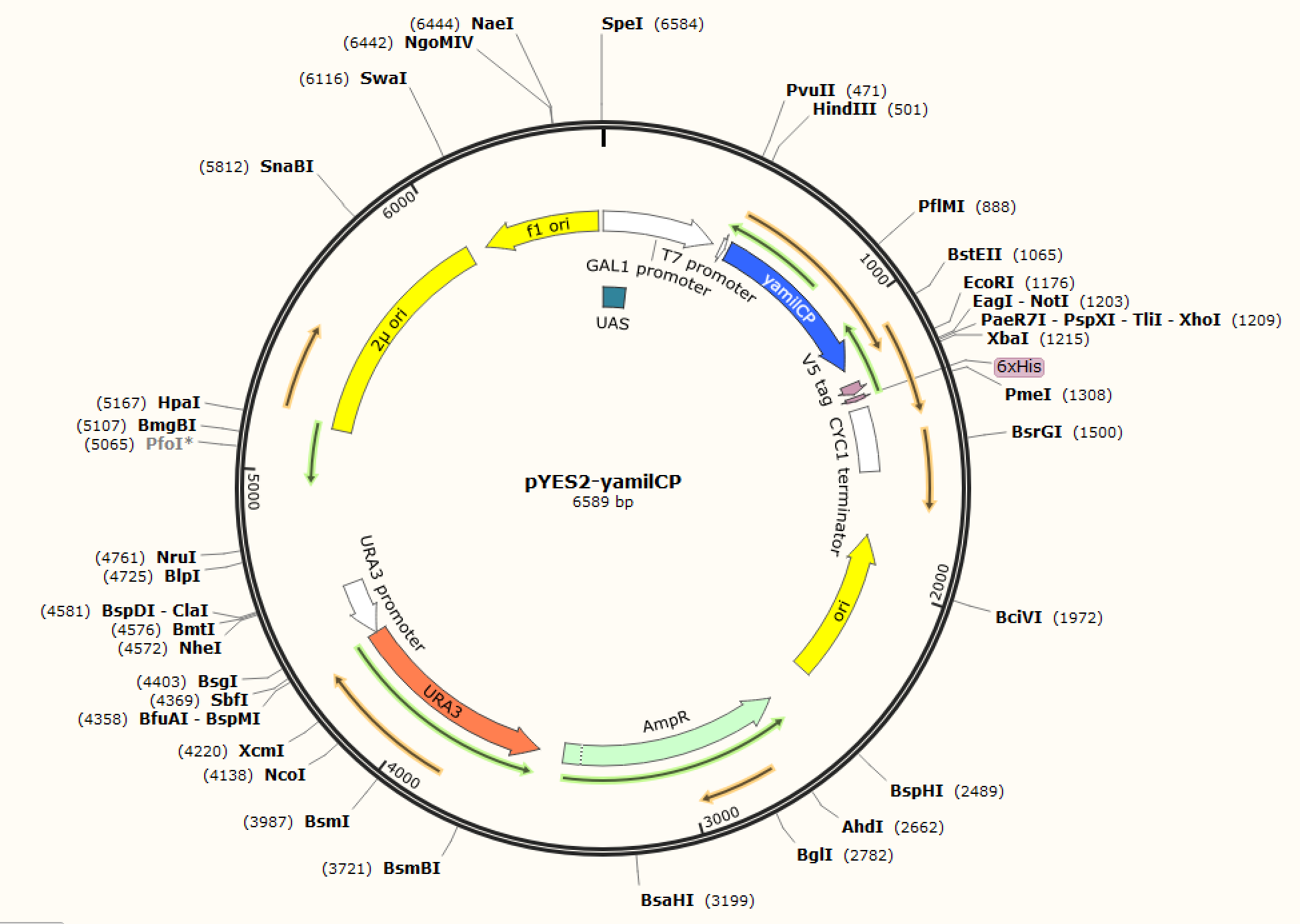
     <p class="left"><b>Figure. 1 The map of plasmid pYES2-amilCP</b>                      <b>Figure. 2 The map of plasmid pYES2-yamilCP</b></p> |      <p class="left"><b>Figure. 1 The map of plasmid pYES2-amilCP</b>                      <b>Figure. 2 The map of plasmid pYES2-yamilCP</b></p> | ||
| − | <p>We respectively cloned the amilCP and yamilCP on the vector of pYES2-NTA.There are the maps of pYES2-amilCP and pYES2-yamilCP (Figure.1 and Figure.2). And we verified the construction of them by double restriction enzyme digestion(Figure.3) and sequencing.</p> | + | |
| + | <p style="font-size:18px !important;">We respectively cloned the amilCP and yamilCP on the vector of pYES2-NTA.There are the maps of pYES2-amilCP and pYES2-yamilCP (Figure.1 and Figure.2). And we verified the construction of them by double restriction enzyme digestion(Figure.3) and sequencing.</p> | ||
[[File:T--SCU-China--scu-improvement-Figure3.png|600px||center|]] | [[File:T--SCU-China--scu-improvement-Figure3.png|600px||center|]] | ||
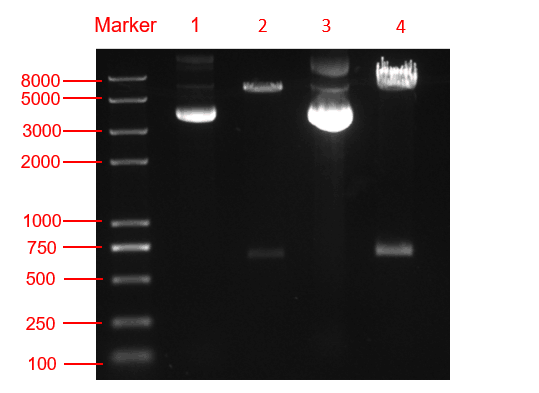
<p class="center"><b>Figure. 3 The double restriction enzyme digestion verification result. lane 1: pYES2-amilCP plasmid, lane 2: pYES2-amilCP plasmid digestion, lane 3: pYES2-yamilCP plasmid, lane4: pYES2-amilCP plasmid digestion, Marker:Trans 2K plus Ⅱ Marker.</b></p> | <p class="center"><b>Figure. 3 The double restriction enzyme digestion verification result. lane 1: pYES2-amilCP plasmid, lane 2: pYES2-amilCP plasmid digestion, lane 3: pYES2-yamilCP plasmid, lane4: pYES2-amilCP plasmid digestion, Marker:Trans 2K plus Ⅱ Marker.</b></p> | ||
| − | <p>Then we transformed pYES2-amilCP and pYES2-yamilCP into <i>S. cerevisiae</i> respectively. Later on, We observed the expression of blue chromoprotein with the induction of galactose (Figure.4).</p> | + | |
| + | <p style="font-size:18px !important;">Then we transformed pYES2-amilCP and pYES2-yamilCP into <i>S. cerevisiae</i> respectively. Later on, We observed the expression of blue chromoprotein with the induction of galactose (Figure.4).</p> | ||
[[File:T--SCU-China--scu-improvement-Figure4.png|600px||center|]] | [[File:T--SCU-China--scu-improvement-Figure4.png|600px||center|]] | ||
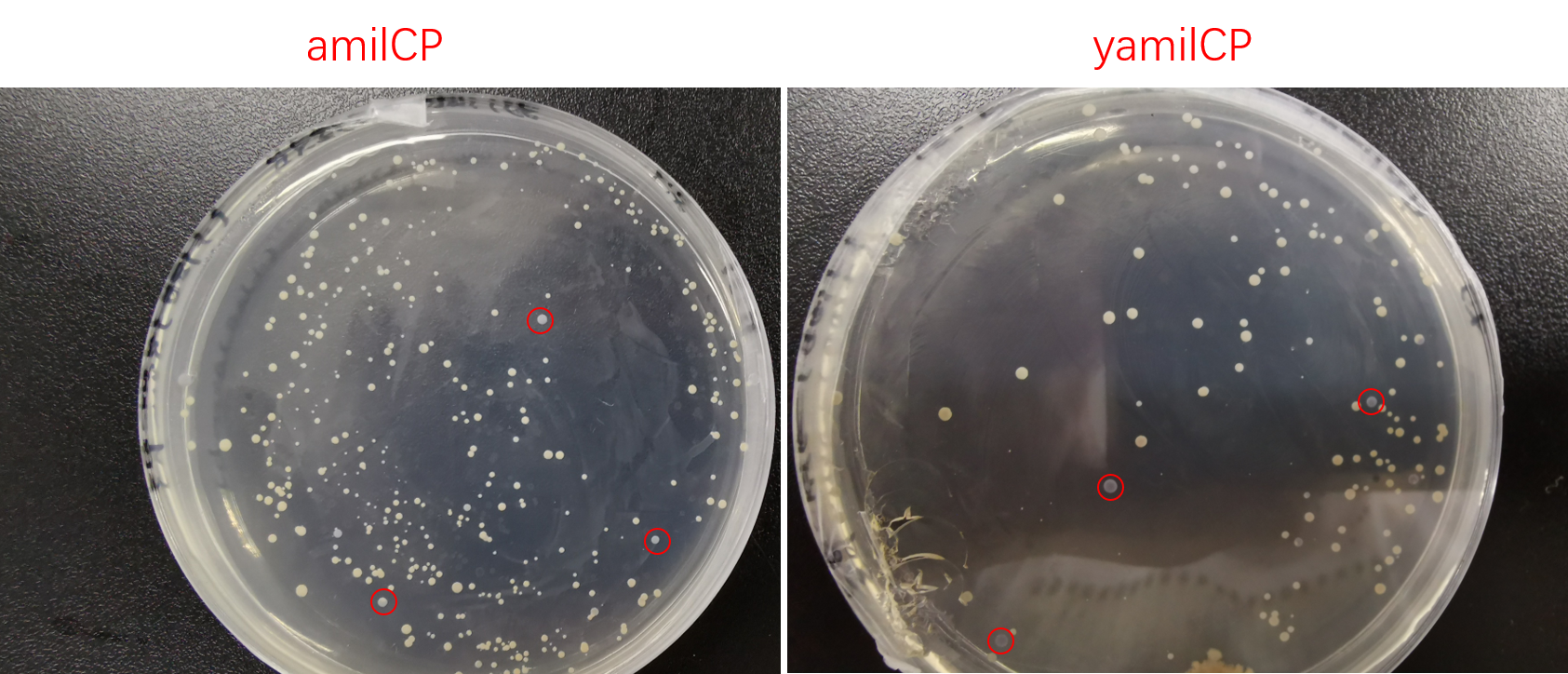
<p class="center"><b>Figure. 4 The colony transformed pYES2-amilCP and pYES2-yamilCP expressed blue chromoprotein.</b></p> | <p class="center"><b>Figure. 4 The colony transformed pYES2-amilCP and pYES2-yamilCP expressed blue chromoprotein.</b></p> | ||
| − | |||
| − | <p>As the result, the improved yamilCP was successfully expressed, and the expression of yamilCP is more quickly than that of amilCP in <i>S. cerevisiae</i>. </p> | + | <p style="font-size:18px !important;">To test the expression effciency of pYES2-amilCP and pYES2-yamilCP in <i>S. cerevisiae</i>, we transformed them into BY4741 and used galactose to induce blue chromoprotein expression. After 10 days, we observed blue colonies on the pYES2-yamilCP plate at first and there was no blue colony on the pYES2-amilCP plate, which confirmed that yamilCP expressed more quickly than amilCP (Figure.5). As Figure. 5, there is no blue clony (0/356) on the amilCP plate while 8 colonies (8/575) on the yamilCP plate. It shows that our improvement is successful.</p> |
| + | |||
| + | |||
| + | <p style="font-size:18px !important;">As the result, the improved yamilCP was successfully expressed, and the expression of yamilCP is more quickly than that of amilCP in <i>S. cerevisiae</i>. </p> | ||
[[File:T--SCU-China--scu-improvement-Figure5.png|600px||center|]] | [[File:T--SCU-China--scu-improvement-Figure5.png|600px||center|]] | ||
<p class="center"><b>Figure. 5 The transformation results of pYES2-amilCP and pYES2-yamilCP into <i>S. cerevisiae</i> BY4741 with galactose induction after 10 days.</b></p> | <p class="center"><b>Figure. 5 The transformation results of pYES2-amilCP and pYES2-yamilCP into <i>S. cerevisiae</i> BY4741 with galactose induction after 10 days.</b></p> | ||
Revision as of 12:49, 20 October 2019
YamilCP is a CDS of blue chromoprotein in S. cerevisiae.
YamilCP is a CDS of chromoprotein which can exhibit strong blue color in S. cerevisiae. It was optimized for S. cerevisiae on the basis of amilCP(BBa_K592009) which is encouraged by Team Uppsala-Sweden 2011. It acts as a reporter in our project to determine the function of Gal1 promoter (pGAL1) and Met3 promoter (pMet3).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
iGEM19_SCU-China:
The yamilCP was improved from amilCP (Part: BBa_K592009), which is a blue chromoprotein gene from Acropora millepora submitted by Team Uppsala-Sweden 2011.
We expected amilCP to be expressed efficiently in S. cerevisiae, so we chose amilCP to be improved. We processed codon optimization on original amilCP sequence and added a consensus sequence (AAAAAA) for S. cerevisiae expression. And the new sequence was named yamilCP.
Figure. 1 The map of plasmid pYES2-amilCP Figure. 2 The map of plasmid pYES2-yamilCP
We respectively cloned the amilCP and yamilCP on the vector of pYES2-NTA.There are the maps of pYES2-amilCP and pYES2-yamilCP (Figure.1 and Figure.2). And we verified the construction of them by double restriction enzyme digestion(Figure.3) and sequencing.
Figure. 3 The double restriction enzyme digestion verification result. lane 1: pYES2-amilCP plasmid, lane 2: pYES2-amilCP plasmid digestion, lane 3: pYES2-yamilCP plasmid, lane4: pYES2-amilCP plasmid digestion, Marker:Trans 2K plus Ⅱ Marker.
Then we transformed pYES2-amilCP and pYES2-yamilCP into S. cerevisiae respectively. Later on, We observed the expression of blue chromoprotein with the induction of galactose (Figure.4).
Figure. 4 The colony transformed pYES2-amilCP and pYES2-yamilCP expressed blue chromoprotein.
To test the expression effciency of pYES2-amilCP and pYES2-yamilCP in S. cerevisiae, we transformed them into BY4741 and used galactose to induce blue chromoprotein expression. After 10 days, we observed blue colonies on the pYES2-yamilCP plate at first and there was no blue colony on the pYES2-amilCP plate, which confirmed that yamilCP expressed more quickly than amilCP (Figure.5). As Figure. 5, there is no blue clony (0/356) on the amilCP plate while 8 colonies (8/575) on the yamilCP plate. It shows that our improvement is successful.
As the result, the improved yamilCP was successfully expressed, and the expression of yamilCP is more quickly than that of amilCP in S. cerevisiae.
Figure. 5 The transformation results of pYES2-amilCP and pYES2-yamilCP into S. cerevisiae BY4741 with galactose induction after 10 days.