Difference between revisions of "Part:BBa K3182100"
| Line 8: | Line 8: | ||
<partinfo>BBa_K3182100 short</partinfo> | <partinfo>BBa_K3182100 short</partinfo> | ||
| + | This part is mainly about the pink-white screening method. For more information and characterization of the carbohydrate binding domain (CBDcipA) please see: <partinfo>BBa_K3182108/partinfo>. | ||
| + | |||
[[File:T--Linkoping_Sweden--fusionproteinillustration.jpg|420px|thumb|right|<b>Figure 1.</b> Mechanism of action for Novosite. The CBDcipA-fusion is attached to a polysaccaride material. By adding thrombin from any source the fusion protein will be cleaved and the C-terminal fusion protein will be released into the solution. By changing the fusion protein to an antimicrobial peptide/enzyme, and using the material as a bandage, the peptide/enzyme can be released into a wound by native human thrombin.]] | [[File:T--Linkoping_Sweden--fusionproteinillustration.jpg|420px|thumb|right|<b>Figure 1.</b> Mechanism of action for Novosite. The CBDcipA-fusion is attached to a polysaccaride material. By adding thrombin from any source the fusion protein will be cleaved and the C-terminal fusion protein will be released into the solution. By changing the fusion protein to an antimicrobial peptide/enzyme, and using the material as a bandage, the peptide/enzyme can be released into a wound by native human thrombin.]] | ||
| Line 46: | Line 48: | ||
<h2>Examples of biobricks assembled with this method</h2> | <h2>Examples of biobricks assembled with this method</h2> | ||
<b>This method was used to assemble</b>: <partinfo>BBa_K3182103</partinfo>, <partinfo>BBa_K3182104</partinfo>, <partinfo>BBa_K3182105</partinfo>, <partinfo>BBa_K3182106</partinfo>, <partinfo>BBa_K3182107</partinfo>, <partinfo>BBa_K3182108</partinfo> into pSB1C3. However, as we quickly noticed, pSB1C3 was not able to replicate in <i>Vibrio natriegens</i> (Vmax). After countless failed transformation attempts, all the above parts were also assembled into a modified pUC19 vector using the pink-white screening method. The pUC19 variants were transformed without any problems into <i>V. natriegens</i> (Vmax). A total of 13 fusion proteins to the CBD was successfully ligated using this method. | <b>This method was used to assemble</b>: <partinfo>BBa_K3182103</partinfo>, <partinfo>BBa_K3182104</partinfo>, <partinfo>BBa_K3182105</partinfo>, <partinfo>BBa_K3182106</partinfo>, <partinfo>BBa_K3182107</partinfo>, <partinfo>BBa_K3182108</partinfo> into pSB1C3. However, as we quickly noticed, pSB1C3 was not able to replicate in <i>Vibrio natriegens</i> (Vmax). After countless failed transformation attempts, all the above parts were also assembled into a modified pUC19 vector using the pink-white screening method. The pUC19 variants were transformed without any problems into <i>V. natriegens</i> (Vmax). A total of 13 fusion proteins to the CBD was successfully ligated using this method. | ||
| − | |||
Revision as of 13:20, 17 October 2019
Contents
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 592
Illegal NheI site found at 615 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 580
- 23INCOMPATIBLE WITH RFC[23]Unknown
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
pT7-CBDcipA-pCons-AsPink This part is mainly about the pink-white screening method. For more information and characterization of the carbohydrate binding domain (CBDcipA) please see: No part name specified with partinfo tag.), seen in Figure 3, as well as a 5'-UTR (BBa_K1758100) region which has been shown to further increase expression in Escherichia coli (E. coli) (BBa_K1758106), ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]).

Theoretical usage of this part
This part utilizes a pCons-AsPink dropout enabling colour-screening for positive colonies. Using BamHI and PstI or SpeI on both this part assembled in pSB1C3 (or vector of choice) and the insert of choice will yield a fusion protein between CBDcipA and the insert. The fusion protein can later be cleaved with thrombin to yield two separate proteins. The C-terminal fusion will have one glycine and one serine added to the N-terminal of the protein.
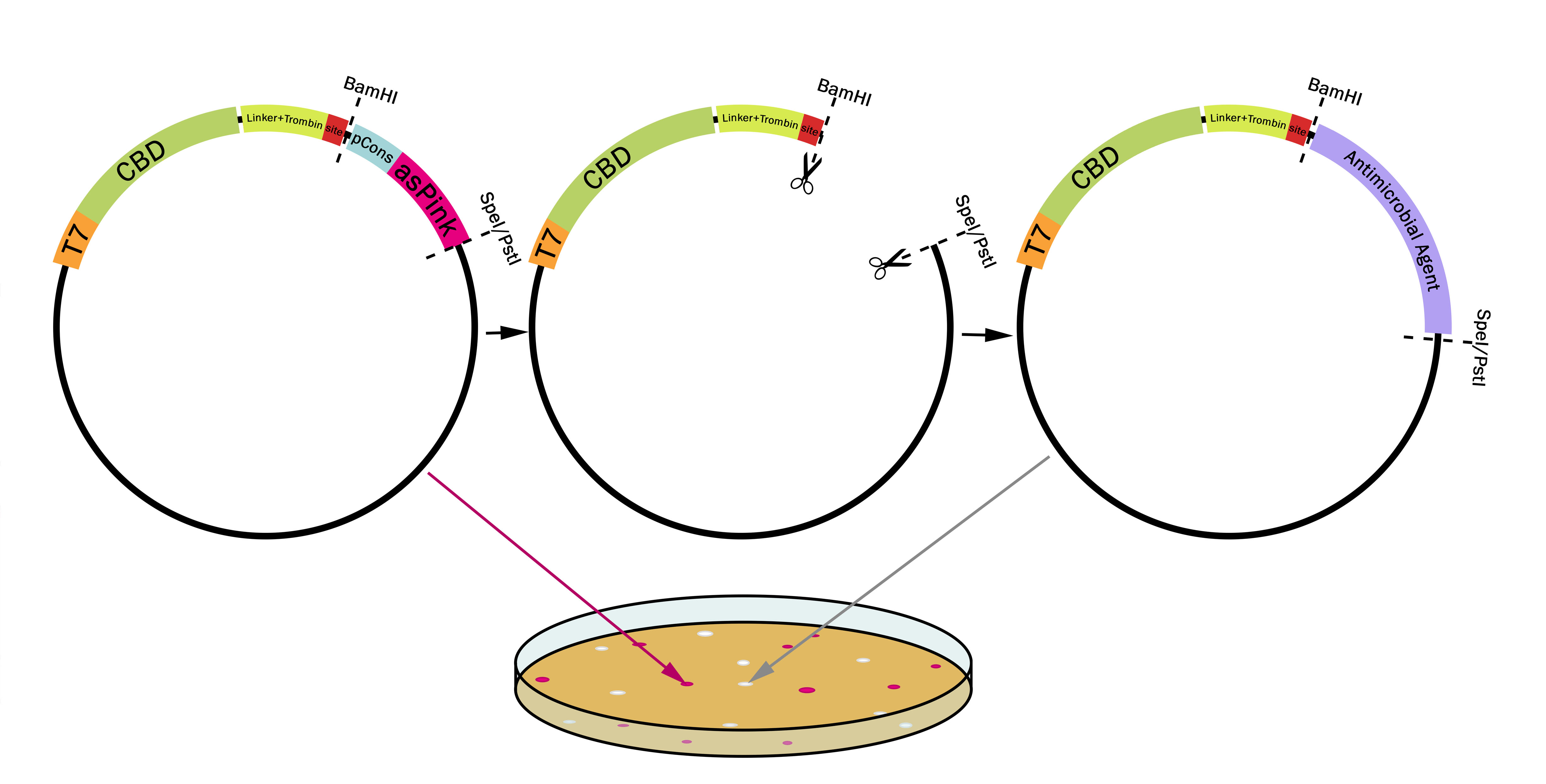
Examples of biobricks assembled with this method
This method was used to assemble: BBa_K3182103, BBa_K3182104, BBa_K3182105, BBa_K3182106, BBa_K3182107, BBa_K3182108 into pSB1C3. However, as we quickly noticed, pSB1C3 was not able to replicate in Vibrio natriegens (Vmax). After countless failed transformation attempts, all the above parts were also assembled into a modified pUC19 vector using the pink-white screening method. The pUC19 variants were transformed without any problems into V. natriegens (Vmax). A total of 13 fusion proteins to the CBD was successfully ligated using this method.
Usage and Biology
Using Pink-White screening
Design any gene which you want to be fused to the CBDcipA with a BamHI recognition sequence in the 5'-end. The biobrick suffix can be used in the 3'-end. Cut the vector and insert with BamHI and PstI (SpeI also works), remove enzymes and mix, no need for gel purification. Using very high molar ratios might not yield any pink colonies at all, a molar ratio insert to vector of 7-20:1 will yield some pink colonies. Transform the host (BL21 (DE3) for quickest results) and incubate at 37 °C overnight, if the color is weak or can not be seen, incubate in 24-37 °C for an additional 16-24 hours.

Results using Pink-white screening
Colonies from the pink-white screening were later sent for sequencing, where all white colonies came back as correctly ligated plasmids. The only colony-screened assembly attempt can be seen in Figure 5C, where all colonies screened showed the correct fragments, as well as all non-pink colonies showing green fluorescence (BBa_K3182108, sfGFP). After the results from Figure 5C had been sequenced, no more colony screenings had to be done. All white colonies sent to sequencing contained the correctly assembled fusion proteins.
