Difference between revisions of "Part:BBa K1982006"
| (One intermediate revision by the same user not shown) | |||
| Line 42: | Line 42: | ||
| − | |||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 52: | Line 50: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1982006 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1982006 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | [1] Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 46, 311-39 | ||
| + | |||
| + | [2] Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121), 823-6 | ||
| + | |||
| + | [3] Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 31(3), 233-9 | ||
| + | |||
| + | [4] Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23 | ||
| + | |||
| + | [5] Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5), 1173-83 | ||
| + | |||
| + | [6] Lauren R. Polstein1 and Charles A. Gersbach. (2015). A light-inducible CRISPR/Cas9 system for control of endogenous gene activation. Nat Chem Biol 11(3): 198–200 | ||
Latest revision as of 13:54, 15 October 2016
tCas9-Vp64(Prokaryotic)
| RBS-tCas9-VP64-HA-FLAG | |
|---|---|
| Function | gene activation |
| Use in | Prokaryotic cells |
| RFC standard | RFC 10 |
| Backbone | pSB1C3 |
| Submitted by | [http://2016.igem.org/Team:NEU-China NEU-China 2016] |
The CRY2/CIBN interaction is entirely genetically encoded. The binding reverses within minutes in the dark, allowing rapid shutoff of transcription by placing samples in the dark. This fusion protein is for use in LACE(light-activated CRISPR/Cas9 effector) system, and a tCas9 fused to its N terminus. To regulate DNA transcription by blue light, the system is based on CRY2/CIBN interaction in which a light-mediated protein interaction brings together two protein (tCas9 and an activation domain VP64) . If we remove the stimulation of blue light, dark reversion of CRY2 will dissociate the interaction with CIBN and shut off transcription.

|
| Figure 1: Construct design. tCas9 can be tagged with transcriptional activators, and targeting these dCas9 fusion proteins to the promoter region results in robust transcription activation of downstream target genes. This tCas9-based activators is the case that tCas9 fused directly to a single transcriptional activator( VP64). |
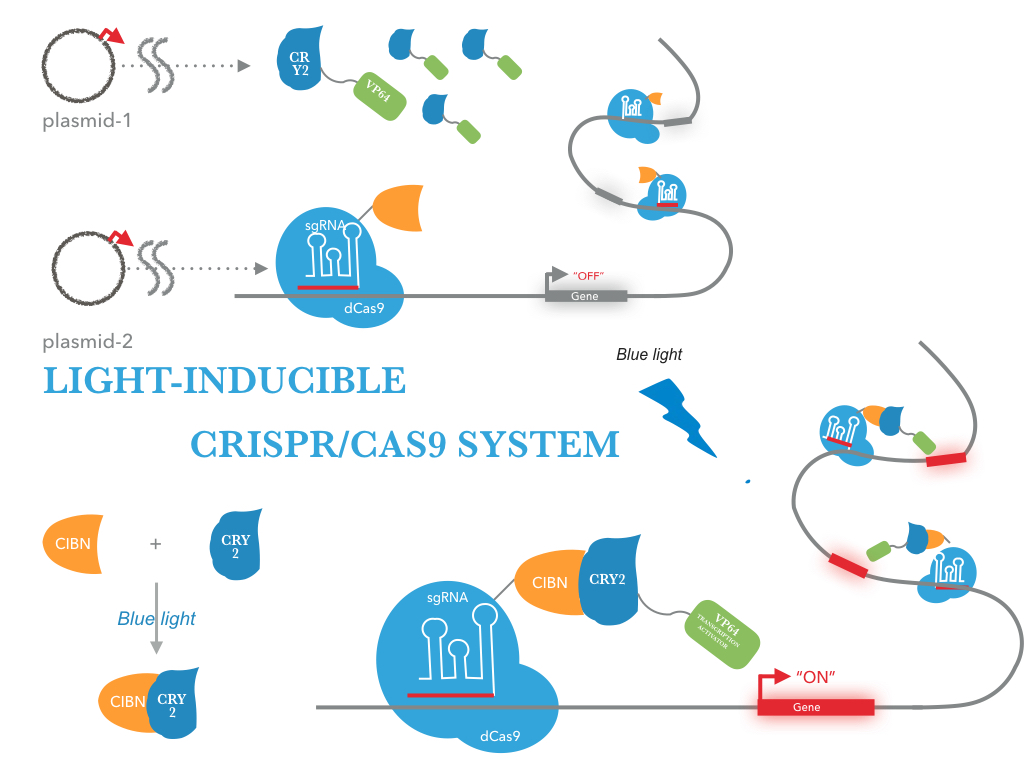
|
| Figure 2: Figure 1 illustrates the detailed design of LACE device |
Usage and Biology
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 10: (underlined part encodes the protein) ATGGACAAG ... GACGACAAATAATAA ORF from nucleotide position 1 to 4344 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2758
Illegal NgoMIV site found at 3667 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 3786
Illegal SapI.rc site found at 1177
Illegal SapI.rc site found at 1419
[1] Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 46, 311-39
[2] Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121), 823-6
[3] Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 31(3), 233-9
[4] Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23
[5] Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5), 1173-83
[6] Lauren R. Polstein1 and Charles A. Gersbach. (2015). A light-inducible CRISPR/Cas9 system for control of endogenous gene activation. Nat Chem Biol 11(3): 198–200
