Difference between revisions of "Part:BBa K1067007"
(→Characterization) |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1067007 short</partinfo> | <partinfo>BBa_K1067007 short</partinfo> | ||
| − | + | Non leaky pBAD promoter with araC constitutively expressed. This biobrick is the same as [[part:BBa_K808000]] but with a change in pBAD promoter and another sequence annotation. The promoter is taken from the [[pBAD SPL]] col. 10 from [http://2013.igem.org/Team:DTU-Denmark 2013 DTU iGEM team]. | |
| + | ===Characterization=== | ||
| + | |||
| + | [[File:690px-Induced vs basal.png|thumb|upright=3|center|Non-induced activity vs inducted activity for a variety of promoters built from the pBAD_SPL. This biobrick (BBa_K1067007) is built with colony 10. The diagonal line represents 1:1, on which the constitutive reference promoter sits. The horizontal blue line represents the induced activity of BBa_K808000.]] | ||
| + | |||
| + | The promoter for this biobrick was taken from a [https://parts.igem.org/PBAD_SPL synthetic promoter library] built to vary the expression and leakiness of [[Part:BBa_K808000]]. The colonies that we selected all show less activity than the the constitutive promoter, and when induced, show higher activity than the constitutive promoter. The promoter taken for this part shows high induced activity and low basal (non-induced) activity. For detail see the graph above. This makes it ideal for expression of lethal proteins and to tightly control expression. | ||
| + | |||
| + | For further characterization and information see the [https://parts.igem.org/PBAD_SPL pBAD SPL page] or go to the 2013 DTU [http://2013.igem.org/Team:DTU-Denmark/pBAD_SPL wiki]. | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | *Expression of lethal proteins | ||
| + | *Tight control | ||
| + | *Direct proteins to the periplasm, see [[Part:BBa_K1067009]] | ||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1067007 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1067007 SequenceAndFeatures</partinfo> | ||
Latest revision as of 12:58, 12 October 2013
Tight pBAD with araC
Non leaky pBAD promoter with araC constitutively expressed. This biobrick is the same as part:BBa_K808000 but with a change in pBAD promoter and another sequence annotation. The promoter is taken from the pBAD SPL col. 10 from [http://2013.igem.org/Team:DTU-Denmark 2013 DTU iGEM team].
Characterization
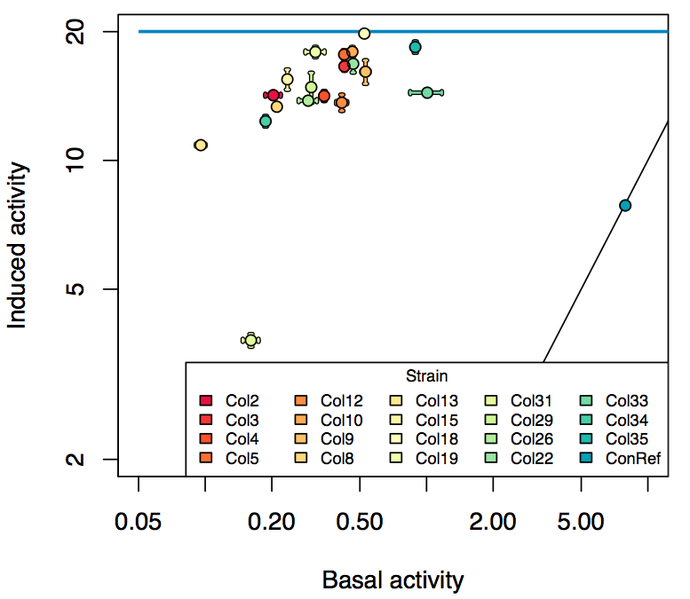
The promoter for this biobrick was taken from a synthetic promoter library built to vary the expression and leakiness of Part:BBa_K808000. The colonies that we selected all show less activity than the the constitutive promoter, and when induced, show higher activity than the constitutive promoter. The promoter taken for this part shows high induced activity and low basal (non-induced) activity. For detail see the graph above. This makes it ideal for expression of lethal proteins and to tightly control expression.
For further characterization and information see the pBAD SPL page or go to the 2013 DTU [http://2013.igem.org/Team:DTU-Denmark/pBAD_SPL wiki].
Usage and Biology
- Expression of lethal proteins
- Tight control
- Direct proteins to the periplasm, see Part:BBa_K1067009
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 1182
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 1182
- 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 1182
Illegal AgeI site found at 979 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
