Difference between revisions of "Part:BBa K792012"
Manugimenez (Talk | contribs) |
Manugimenez (Talk | contribs) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
The high level structure of the device is: | The high level structure of the device is: | ||
| − | * '''''BamHI''''' restriction site ''(introduced | + | * '''''BamHI''''' restriction site ''(introduced to facilitate cloning into yeast expression vectors (pEG202, pCM180 series) - '''this is not essential''' for the device function)'' |
* '''''Kozak''''' consensus sequence for initiation of translation | * '''''Kozak''''' consensus sequence for initiation of translation | ||
* '''''Signal''''' peptide that targets the product of the gene for secretion | * '''''Signal''''' peptide that targets the product of the gene for secretion | ||
| Line 18: | Line 18: | ||
| − | {| style="width:100% | + | {| style="width:100%" |
|- | |- | ||
| − | | style="background: red | + | | style="background: red; width:11%" | '''Important note: ''' |
| − | | style="background: #D8D8D8 | + | | style="background: #D8D8D8" | this part has been submitted to the registry using a direct synthesis DNA sample, so it does not contain scars from ''BB assembly standard''. |
|} | |} | ||
| − | {| style="width:100% | + | {| style="width:100%" |
| + | |- | ||
| + | | style="background: yellow; width:11%" rowspan="2"| '''See also: ''' | ||
| + | | style="background: #D8D8D8" | Another '''''Trp-rich export and enhanced import''''' composite device [[Part:BBa_K792010]]. | ||
| + | |- | ||
| + | | style="background: #D8D8D8" | Similar design '''''His-rich export and enhanced import''''' composite device [[Part:BBa_K792011]]. | ||
| + | |} | ||
| + | |||
| + | {| style="width:100%; background: #FFBF00" | ||
| + | |- | ||
| + | | [[Image:BsAs2012-Biohazard.png | 70px]] | ||
| + | | This device contains <partinfo>K792004</partinfo>, a trojan peptide. Extreme your safety measures when dealing with strains containing this part: '''avoid direct contact with them or conditioned medium and discard them as pathogenic residues.''' | ||
| + | |} | ||
| + | |||
| + | {| style="width:100%; background: #CEE3F6" | ||
|- | |- | ||
|The device was originally designed and built to be used in [http://2012.igem.org/Team:Buenos_Aires iGEM BsAs 2012 project] to implement a ''cross-feeding'' regulatory system between two different yeast strains with specific auxotrophy. [http://2012.igem.org/Team:Buenos_Aires Visit our wiki] to read details about the designing process and implementation details. Also feel free to contact us for any question regarding this part. | |The device was originally designed and built to be used in [http://2012.igem.org/Team:Buenos_Aires iGEM BsAs 2012 project] to implement a ''cross-feeding'' regulatory system between two different yeast strains with specific auxotrophy. [http://2012.igem.org/Team:Buenos_Aires Visit our wiki] to read details about the designing process and implementation details. Also feel free to contact us for any question regarding this part. | ||
| Line 30: | Line 44: | ||
|} | |} | ||
| + | == Data == | ||
| + | |||
| + | {| | ||
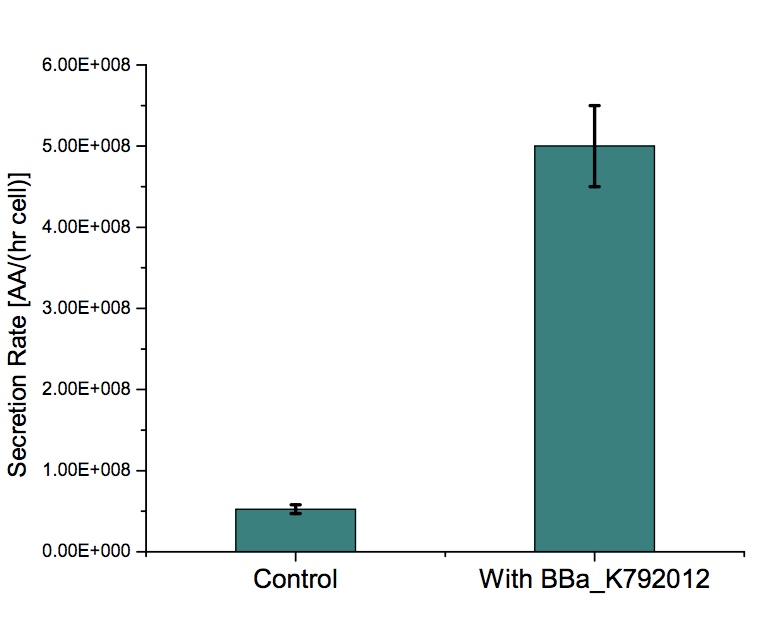
| + | |To determined the devices Tryptophan's secretion rate we used a [http://2012.igem.org/Team:Buenos_Aires/Results/DevicesTesting#Secretion_Rate_of_Trp_as_a_function_of_culture_growth simple model] that describes the behaviour of a strain transformed with the device that produces and secretes this amino acid and how it accumulates in the medium. The secretion rate is the ratio between the concentration of tryptophan at a given time and the time integral of the culture density. | ||
| + | |||
| + | |||
| + | * We measured the amount of tryptophan in the medium acquiring the florescence spectra of the medium and compering with a calibration curve. | ||
| + | * We used ''W303a yeast'' strains, and ''pCM182 plasmid''. | ||
| + | * To take into account aminoacid natural diffusion through cell's membrane, we used a strain transformed with the pCM182 plasmid but without BBa K792010 device, as a control. | ||
| + | |||
| + | |||
| + | As shown in the figure, the tryptophan '''secretion rate in the strain with the device is almost 10x higher''' than the control. | ||
| + | More information about experiments we performed with this device can be found in [http://2012.igem.org/Team:Buenos_Aires/Results/DevicesTesting BsAs 2012 wiki]. | ||
| + | |||
| + | |||
| + | | | ||
| + | {| class="wikitable" | ||
| + | | [[Image:BsAs2012-result-K792012.jpg | 500px]] | ||
| + | |} | ||
| + | |} | ||
Latest revision as of 23:33, 27 October 2012
Yeast exportable Trp-rich peptide w/enhanced import (2)
This composite device produces and exports a Tryptophan rich peptide, that is import enhanced (thanks to a trojan peptide). This device is intended to be used with Yeast chassis, so it is composed by subparts designed also to be use in yeast.
The high level structure of the device is:
- BamHI restriction site (introduced to facilitate cloning into yeast expression vectors (pEG202, pCM180 series) - this is not essential for the device function)
- Kozak consensus sequence for initiation of translation
- Signal peptide that targets the product of the gene for secretion
- Trojan peptide, to increase internalization in target cell (Polyarginine)
- Payload: this is the exported Tryptophan rich domain of the protein (a custom designed peptide that we called PolyWb)
The device's sequence has inherited HindIII restriction site from the parts it's made of.
| Important note: | this part has been submitted to the registry using a direct synthesis DNA sample, so it does not contain scars from BB assembly standard. |
| See also: | Another Trp-rich export and enhanced import composite device Part:BBa_K792010. |
| Similar design His-rich export and enhanced import composite device Part:BBa_K792011. |

|
This device contains BBa_K792004, a trojan peptide. Extreme your safety measures when dealing with strains containing this part: avoid direct contact with them or conditioned medium and discard them as pathogenic residues. |
Data
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]