Part:BBa_K3122005
Constitutive medium promoter + strong RBS + AEGIS's lamB display system + Terminator
This composite part consists of our AEGIS's lamB display system for Gram- bacteria, expressed under constitutive promoter and strong RBS regulation, and a terminator. It was created using a MoClo single-pot reaction, and assembled into pARK1 plasmid in the same step.
This transcriptional unit includes the following parts:
- BBa_K3122003: Promoter J23107 adapted to type IIS assembly
- BBa_K2656009: RBS B0030 adapted to type IIS assembly
- BBa_K3122000: Coding sequence. LamB coding sequence. This part is our AEGIS' lamB display system.
- BBa_K2656026: Terminator B0015 adapted to type IIS assembly
To see the design of this part go to the Design tab.
Description
As explained in BBa_K3122000, E. coli LamB is one of the best characterized and most flexible heterologous peptide carriers, and can tolerate insertions of peptide tags between amino acid residues 153 and 154 (Sousa, 1996). Our group have adapted this functional display system to type IIS enzymes grammar, and called it AEGIS's lamB display system for Gram-negative bacteria.
It has been used as negative control for histidine-related assays in our SELEX procedure, but the main purpose of this part is to prove tan our display system MoClo adapted is functional on its own, before expressing any tag.
We then decided to express a six-unit histidine tag in our LamB display system, conforming the transcriptional unit BBa_K3122004.
Characterization
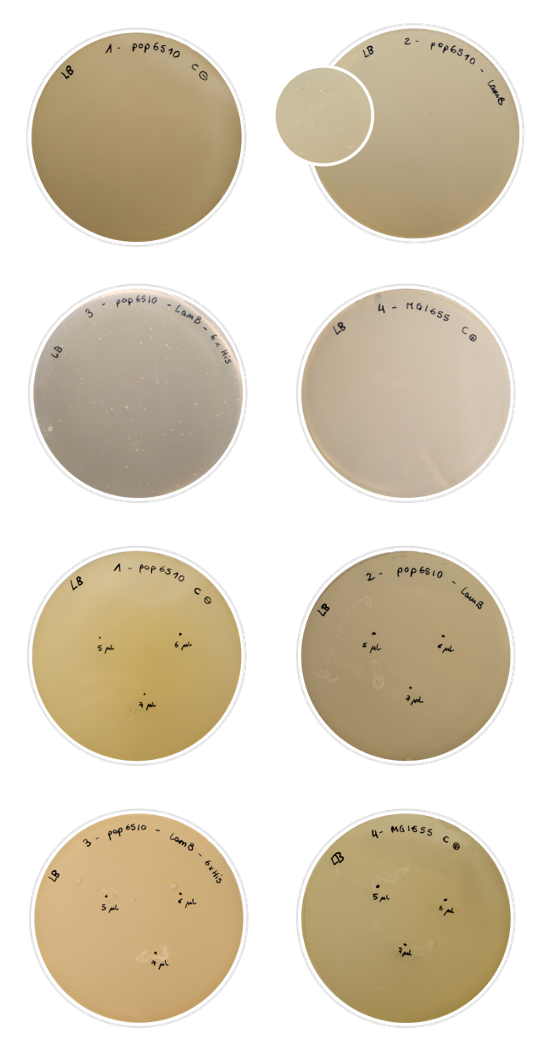
For this characterization, we chose to perform a rather simple test that nonetheless gives conclusive results: the Lambda phage assay. The Lambda phage is a bacteriophage which infects the cell using the LamB protein. We used pop6510 cell line, which does not express LamB. Therefore, if there is no expression of our constructs, the phage cannot infect the cell and there will not be any lysis. On the other hand, if the Lamb display system is expressed properly, the phage will be able to infect the cell, and lysis spots shall appear in the Petri dish.
In this experiment, we characterized the expression in the outer membrane of the LamB protein both with the 6xHis tag and without. We aimed to tell if we had achieved a correct expression of the protein, and if the addition of the His-tag into the permissive loop would compromise this expression.
To prepare the test, we first dabbed the white colonies and made a liquid inoculum. The grown inoculum was treated as explained in the Fague lysis protocol, and then we conducted two different assays:
- Plating in LB agar plates a mix of soft-agar and the result of an incubation of 100μl of the liquid inoculum with 100 μl of phage.
- Plating soft-agar with the bacteria inoculum in LB agar plates with known droplets of Lambda phage: 5,6 and 7 μl in three punctual locations in the Petri dish.
We used regular MG1655 E. coli cells (expressing LamB normally) as positive control, and our pop6510 cells as negative control. The results obtained were positive:
- In the first assay, negative control plate was covered with a bed of cells and positive control plate was clean of cells, as expected. About problem samples, we could observe very few colonies in the petri dish, meaning that the majority of the cells were expressing LamB and therefore lysis has occurred in a high percentage of cells. In the second assay, similar good results were obtained: bald patches appeared in the zone where the Lambda phage was inoculated in the case of problem samples and positive control, while negative control had grown generously.
- In conclusion, the assays could be confirmed as successful. The results explained above were very similar in both constructions, confirming that the introduction of the Tag does not affect the correct expression of LamB in the E. coli outer membrane. However, this result should not be extrapolated to every tag. Bigger or longer tags may lead to misleading results, making this assay unuseful to check its presence.
How to use it
This is the creation of our AEGIS’s LamB display system that we created. It can be used by future iGEM teams on their MoClo reactions with the desired tag to create level 1 constructs. The only requirement is to adapt the tag sequence to the new scars designed.
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 11
Illegal NheI site found at 34 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 212
Illegal AgeI site found at 524
Illegal AgeI site found at 1289 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 527
Illegal BsaI.rc site found at 521
Judging
Even if several different display systems exist on the iGEM repository, AEGIS's lamB display system for Gram- bacteria stands out among them. The advantages of this approach are notable: not only can it be used for bacteria with cell wall ensuring its right functioning, but also it is really simple and easy to use following MoClo standard.
This transcriptional unit, together with BBa_K3122004, has been used to characterize the existing BBa_J23107 Promoter basic part (adapted to type IIS gramar in the new part BBa_K3122003), BBa_K2656009 RBS basic part and BBa_K2656026 Terminator basic part.
References
C. Sousa, A. Cebolla and V. de Lorenzo, "Enhanced metalloadsorption of bacterial cells displaying poly-His peptides", Nature Biotechnology, vol. 14, no. 8, pp. 1017-1020, 1996. Available: 10.1038/nbt0896-1017 [Accessed 20 October 2019].
| None |