Part:BBa_K1170003
p-atp2 in F0F1 ATPase operon from C. glutamicum ATCC 13032
Alkali inducible promoter from the F0F1 operon in C. glutamicum ATCC 13032. This can be used to generate a response from a biological system based on high pH.
Team CAU_China shortened this part to include only the -35 box, -10 box, and TSS of Patp2(BBa_K3796204).
Usage and Biology
Team BIT-China 2015 improved this part by making it compatible with RFC10 (see: BBa_K1675022).
BBa_K1675022 is the recombinant of P-atp2 mutant, B0034 and LacZ alpha
Contribution From CAU_China 2021
Group: CAU_China,2021 https://2021.igem.org/Team:CAU_China
Author: Han hanwen & Wang Fengyi
Summary: Added information about how this promoter responds to pH stress and specified its features; Shortened its sequence and characterized how it works in Corynebacterium glutamicum
Usage
As an inducible promoter, Patp2 can be used to satisfy the needs for pH responses in project design. For example, it can be used for specifically expressing the target protein in alkaline environment, in which the target protein is often related to the tolerance of high pH environment, like a proton transporter[2]. Also, it can be used together with other pH inducible promoter for a system that can respond and adapt to different pH, from acidic to alkaline.
In our project, CAU_China 2021 used this part in their composite kill switch to make sure the engineered bacteria would die when the alkalinity of the soil decrease to an expected level.
Biology
P-atp2 is a part of the FoF1-ATPase operon in Corynebacterium glutamicum.
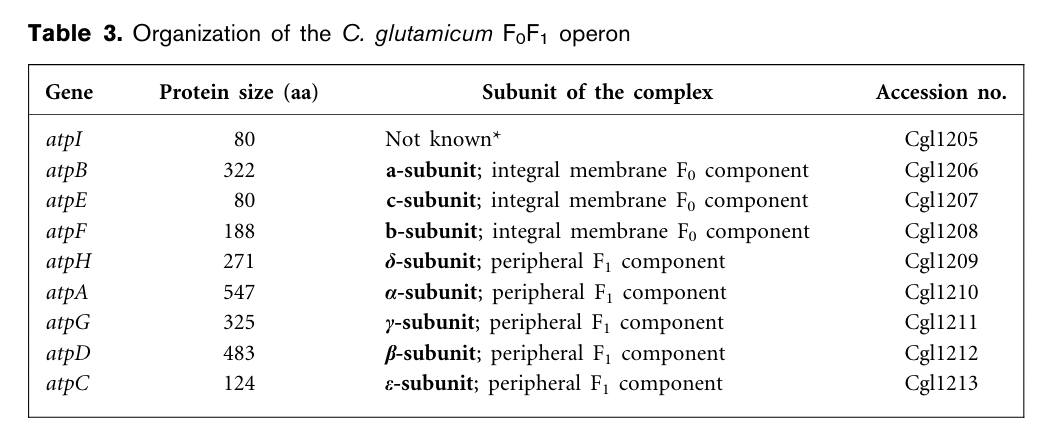
Preliminary studies suggested that C. glutamicum is a moderately alkali-tolerant organism resulted from pH-regulated F0F1 operons which encodes the F0 and F1 multiprotein complexes of the ATP synthase that is involved in the formation of ATP using the electrochemical force of the membrane proton gradient. The genetic organization of the C. glutamicum F0F1 ATP synthase operon maintains the canonical order of the eight structural genes, atpBEFHAGDC.
P-atp2 is the promoter of atpB and its sequence structure is shown in Fig.2. The transcription start points are indicated by bent arrows in the nucleotide sequence. Ribosome-binding sites are shaded in grey, and possible stop codons for the atpI gene are indicated by dashed lines. The -10 and -35 boxes are underlined and in italic letters, and the ATG translation start triplets for atpB, respectively, are shown in bold.[1]
Under alkaline conditions, σH can bind to the P-atp2 promoter of the FoF1-ATPase operon and begin to express downstream genes. When the bacteria grow at alkaline pH, the external pH of the bacteria triggers the change of internal pH, which then acts as an intracellular signal to increase the content of σH factor, so that the binding efficiency of σH with p-atp2 promoter increases and the expression of downstream genes increase as well. Subsequently F0F1 operon expression is induced, thus allowing a higher rate of ATP synthesis and increased growth at its optimal alkaline pH.
Characterization
CAU_China 2021 shortened this part to include only the -35 box, -10 box, and TSS of Patp2(BBa_K3796204) and characterized its function.
We aim to verify that the promoter Patp2 can respond to pH stress in Corynebacterium glutamicum and we also want to document its response towards different alkalinity.
We use the gene circuit below(BBa_K3796207) to test the function of Patp2 by the fluorescence of GFP.
We build the gene circuit by ClonExpress II one-step cloning kit (Vazyme Biotech, China). Particularly, we destroyed the tac promoter on the plasmid pXMJ19 to get rid of its influence.
After sequencing and amplifying, that vector as well as unmodified pXMJ19 were transferred into Corynebacterium glutamicum separately and cultivated on plates. After cultivating them in the LB liquid culture medium at 100 rpm, 30℃ for 12h, the OD600 of the two bacteria were adjusted to nearly the same(nearly 2.0) and inoculated in the pH gradient LB liquid medium. After a cultivation of 24h, the bacterial solution was collected and washed with PBS. Then we measured its fluorescence intensity by HITACHI F-7000, a fluorescence spectrophotometer, according to the excitation light of 488nm and emission light of 507nm and OD600. Data are shown below:
Through two-way ANOVA, we can know that there’s significant difference between the control group and experiment group under different pH conditions. The relative fluorescence intensity of the control group is relatively stable at the pH range of 7-9.5, while there is a significant increase of relative fluorescence intensity in experimental group. Data shows that the difference of relative fluorescence intensity between the two groups remains high between pH=8.5 and pH=9.0, and reaches its peak near pH=9.0, indicating that the promoter Patp2 has the most ability to enhance its downstream gene expression in this pH range. Also, we can see a sharp drop of the relative fluorescence intensity in the experiment group when the pH is more than 9.5. We assume that this is because C.glutamicum can’t tolerate such a high alkalinity stress and the OD600 decreases a lot, making the relative fluorescence intensity seems abnormal or irregular.
Three parallel experiments were done later, which supported our opinions. We finally come to the conclusion that Patp2 can respond to pH changes in C.glutamicum, and high alkalinity can improve the expression of the downstream genes of Patp2. According to our data, the peak occurs near pH=9.0, which happens to fall in the range of the alkalinity of saline-alkaline soil.
References For Contribution From CAU_China
[1]Barriuso-Iglesias,M. , et al"Transcriptional analysis of the F0F1 ATPase operon of Corynebacterium glutamicum ATCC 13032 reveals strong induction by alkaline pH. " Microbiology (2013).
[2] Barriuso-Iglesias, M. , et al. "Transcriptional control of the F0F1 -ATP synthase operon of Corynebacterium glutamicum : SigmaH factor binds to its promoter and regulates its expression at different pH values." Microbial Biotechnology 6.2(2013).
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 625
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 625
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 625
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 625
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 625
Illegal AgeI site found at 30 - 1000COMPATIBLE WITH RFC[1000]
| None |