Plasmids
Plasmids are circular, double-stranded DNA molecules typically containing a few thousand base pairs that replicate within the cell independently of the chromosomal DNA. Plasmid DNA is easily purified from cells, manipulated using common lab techniques and incorporated into cells. Most BioBrick parts in the Registry are maintained and propagated on plasmids. Thus, construction of BioBrick parts, devices and systems usually requires working with plasmids.
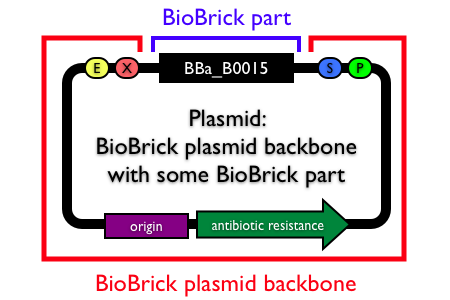
Note: In the Registry, plasmids are made up of two distinct components:
- the BioBrick part, device or system that is located in the BioBrick cloning site, between (and excluding) the BioBrick prefix and suffix.
- the plasmid backbone which propagates the BioBrick part. The plasmid backbone is defined as the sequence beginning with the BioBrick suffix, including the replication origin and antibiotic resistance marker, and ending with the BioBrick prefix. [Note that the plasmid backbone itself can be composed of BioBrick parts.]
Many BioBrick parts in the Registry are maintained on more than one plasmid backbone!
To construct, assemble, or operate BioBrick parts, you are probably looking for a plasmid backbone ... not a plasmid. Please go to the Plasmid backbones collection to see all available plasmid backbones.
However, the Registry does have a small collection of plasmids. These are largely legacy parts and not useful for construction of BioBrick parts, devices and systems. However, we include them here for completeness.
SynBERC Tumor killing bacteria testbed
One of the testbeds for the NSF-funded [http://synberc.org Synthetic Biology Engineering Research Center (SynBERC)] is designing and engineering modules that will be integrated to construct a bacterium capable of moving to and attacking a chemical or biological entity; for example, a tumor or a chemical warfare agent. There are a number of environmental cues that bacteria could use to distinguish a tumor from healthy tissue. The environment is hypoxic and more nutritious, and bacteria grow to significantly higher cell densities (Yu et al., 2003). Components that sense these differences can be linked to genetic circuits that integrate the information. The circuits will activate engineered pathways that control bacterial chemotaxis and the interaction between the bacterium and a mammalian cell. These systems will be engineered into a non-pathogenic E. coli chassis.
| Name | Description | Resistance | Copy Number | Origin | Length | Default Insert | PCR |
|---|---|---|---|---|---|---|---|
| BBa_J61004 | pAC-TetInv | 5559 | |||||
| BBa_J61005 | pAC-LuxInv | 7181 | |||||
| BBa_J61006 | pBACr-AraInv | 10830 | |||||
| BBa_J61008 | pBACr-FdhInv | 9764 | |||||
| BBa_J61009 | pAC-LuxGFP | C | p15A | 5130 | p15A | p15A |
| Chris Anderson, an assistant professor of bioengineering at UC Berekeley and the SynBERC testbed leader, has submitted a set of plasmids associated with his paper on Environmentally Controlled Invasion of Cancer Cells by Engineered Bacteria. Please read the paper for details or contact Chris for details. |
<biblio>
- Anderson pmid=16330045
</biblio>
Other miscellaneous plasmids
The Registry also has miscellaneous other plasmids. These plasmids have not undergone Registry curation, but we include them here for completeness.