Part:BBa_K880001:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K880001
User Reviews
UNIQ3cbf4debc3297ac7-partinfo-00000000-QINU
|
Michigan iGEM 2013 |
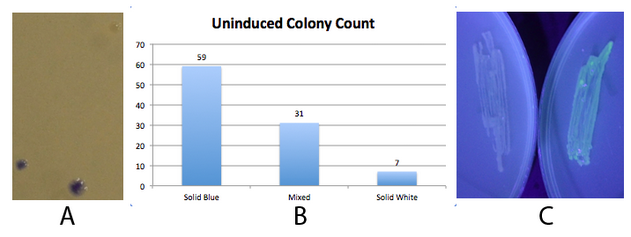
This part did not work very well. DO NOT USE IT!!! In the presence of either hbiF or fimE, it was barely able to flip. Below is characterization of the new fim switch K1077005. Fig. 1 - The fim transcriptor is capable of changing states completely and unidirectionally NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with either constitutive hbiF or fimE and BBa_K1077007 (J23100 fim switch, ON orientation), plated on LB plates and grown overnight for ~16 hours. The state of the switch was assayed by using an asymmetric digest assay on PCR amplified switch. There are two hincII sites located within the K1077007 switch, one of which changes position depending on the state of the switch. The result is that when the switch is in the ON position, a 870bp, 273bp, and 248bp band is produced. When the switch is in the OFF position, a 680bp, 473bp, and 273bp band is produced. A and B)The digest assay was quantified using densitometry and showed greater than 95% of the switch in the desired state (ON when co transformed with constitutive hbiF and OFF when co transformed with constitutive fimE). This is consistent with hbiF’s previously observed functionality of catalyzing the inversion of fimS from the OFF to ON orientation and fime’s previously observed functionality of catalyzing the inversion of fimS from the ON to OFF orientation. C and D) A gel of the digest assay fragments quantified in A and B. The faint ~870bp band in 1D, seemingly indicating residual OFF state, may correspond to the constitutive recombinase generator which is 858bp. We did not have time to cure the bacteria of the constitutive recombinase generator plasmid. Additionally, the switch is on the high copy pSB1C3 plasmid and so it could be that some switch plasmids are escaping recombination. We did not have time to move the switch to a low copy plasmid or the chromosome. Fig 2. - An inducible fim transcriptor system changes states and produces protein output NEB 10-beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were co-transformed with K1077007(amilCP j23100 fim switch) or K1077003(GFP j23100 fim switch), and K1077002 (aTc inducible fimE, HSL inducible hbiF) and plated on to LB plates with or without inducer. A) Close up of 3 colonies on a plate containing K1077007 and K1077002 co transformants. Three distinct phenotypes were observed: Solid Blue colonies(bottom left), Mixed colonies that had distinct white and blue regions(bottom middle), and Solid White colonies(top right). Therefore, the uninduced fim transcriptor in the context of K1077002 is subject to leaky fimE and/or hbiF activity. B) Quantification of the phenotypes observed on the plate in A. C) When induced with 4.32µM aTc and grown overnight (left) , no GFP is produced as expected due to induced fimE turning the switch OFF. When induced with 1µM HSL and grown overnight (right), GFP is produced as expected due to induced hbiF turning the switch ON. Given the absence of GFP in the aTc induced switch shown in 2C, leaky hbiF expression is overcome by the induced fimE expression. Nothing can be said about induced hbiF overcoming leaky fimE since the green may still indicate only some of the plasmids are flipped. Whether or not the phenotypes of the colonies observed in 2A are due to variable leakage rates of each recombinase or variable enzymatic activity of each of the recombinases, or both, can not be determined. Since these are co transformations, the white and mixed colonies observed could simply be due to chance production of fimE only or first in the first generation, flipping the whole population of switch plasmids in the cell (~1). Subsequent generations might inherit the switch mostly in the initial state. We did generate only inducible hbiF and fimE plasmids, but did not have time to co transform them with the switch or submit them to the registry. These would allow us to test the effects of leakage of each recombinase on the switch. Other assays, such as time course data of recombinase activity under the same inducible system, would be needed to determine relative rates of recombinase activity. Moving the inducible recombinase generator to a low copy plasmid, may reduce leakage effects. Ideally, a more tightly regulated inducible system should be coupled to the recombinases. Promoter and/or rbs optimization should be performed to minimize recombinase leakage above the threshold needed for significant flipping activity. |
UNIQ3cbf4debc3297ac7-partinfo-00000002-QINU