Part:BBa_K729006
J23119-RBS-Laccase
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 286
- 1000COMPATIBLE WITH RFC[1000]
Description
The laccase enzyme has been shown to degrade polyethylene when being expressed by a number of different bacteria and fungi. This is due to conserved copper binding sites which couple the oxidation of a substrate with the cleavage of dioxygen bonds, leading to the capability to degrade plastics (particularly polyethylene).
By driving the laccase production using a strong constitutive promoter, we expect overexpression of the protein which will allow it to be secreted into the extracellular medium where it can act upon the target plastic.
Characterisation
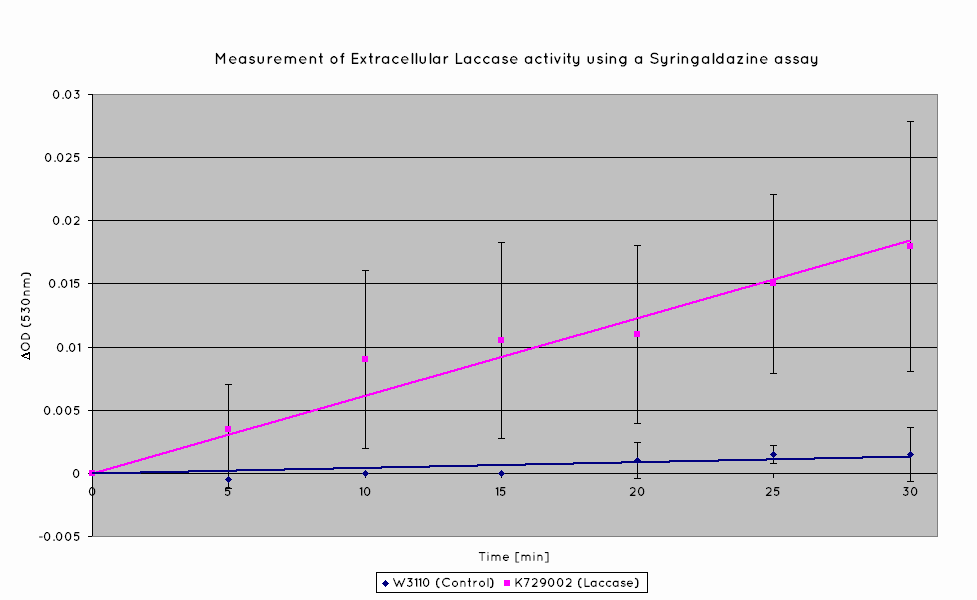
Plastic degradation is mediated via a laccase protein. As such, we will be using an enzymatic activity assay to determine that the laccase enzyme is expressed. Laccase catalyses the oxidation of syringaldazine, a reaction that exhibits an observable OD change at 530nm. A sample of syringaldazine can be used as a blank in a spectrophotometer, against a sample containing syringaldazine and our laccase sample, allowing the rate of oxidation to be measured, and hence the enzymatic activity of laccase.
In order to determine the effectiveness of laccase in degrading plastic, we will expose strips of various types of plastic to the laccase expressing bacteria, before viewing the strips under a scanning electron microscope. This will allow us to compare the pitting in plastic samples treated with laccase, to the untreated samples, allowing us to determine the extent of plastic degradation.
Results
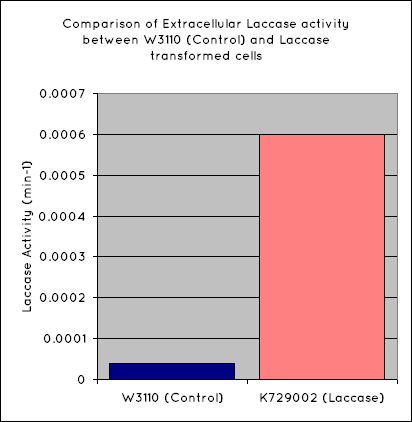
Our results indicate a significantly higher rate of oxidation from the cells containing our BioBrick than the control cell line. This indicates that our transformed E. Coli have successfully produced laccase, and in significant enough quantities that it is released into the extracellular space. This allows it to oxidise the syringaldazine utilised in the laccase assay.
The Scanning Electron Microscope (SEM) images below show the effects of subjecting Low Density Polyethylene (LDPE) to three different sets of conditions in order to visualise the degradative properties of cells containing our Laccase construct (BBa_K729006). We can then also compare this with the ability of untransformed E. coli W3110 and mechanical shear in water to breakdown the LDPE target.
All samples were in suspension at 37°C and 200rpm in an incubated shaker for 3 days.
Low magnification (x100) images were used to observe the macro-trend in zonal enzymatic activity. There appears to be increased ‘roughness’ (shown by the small protrusions on the surface as well as areas of darker shading, which represent areas of greater contour), across the surface of the plastic as we progress from the sample treated with water to untransformed E. coli W3110 to E. coli transformed with the UCL Laccase construct.
High magnification (x10 000) images were taken to give more high resolution information regarding the activity of the enzyme on a much smaller scale. The SEM images highlight increased amounts of ‘pitting’ (the generation of small holes in the plastic due to it’s structural breakdown), as we go from water to E. coli W3110 to bacteria containing BBa_K729006. These pits are shown by the much darker areas. This small scale breakdown has a cumulative effect which results in the overall trend seen at the lower magnification.
In conclusion, this collection of SEM images indicates an ability both of the BBa_K729006 transformed cells to produce the Laccase enzyme at greater titres than untransformed bacteria (though the E. coli W3110 is still seen to have some degradative effect), and of the Laccase produced to degrade the polyethylene target.
| Water | W3110 | Laccase | |
|---|---|---|---|
| Low Magnification (100x) | 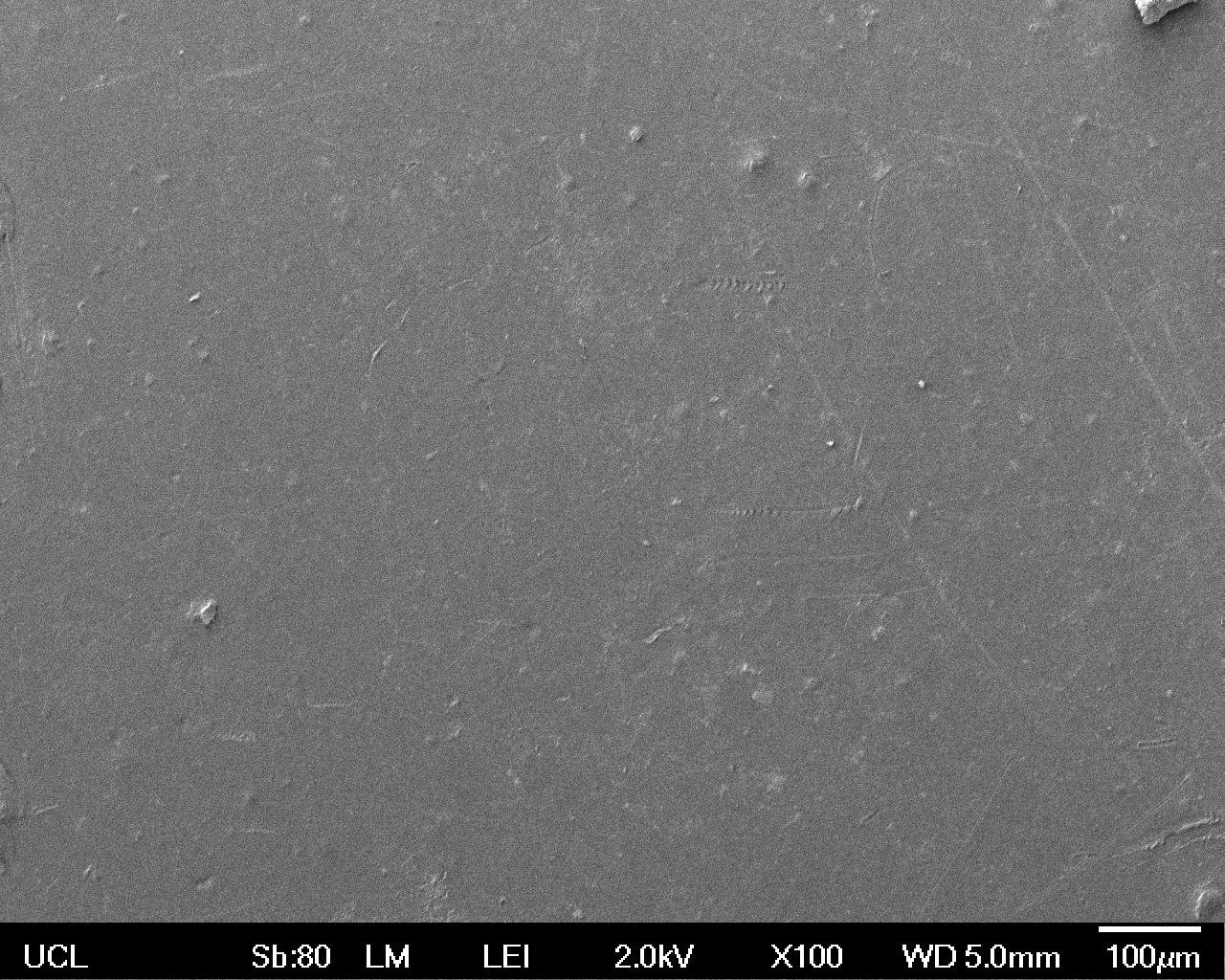 |
 |

|
| High Magnification (10 000x) | 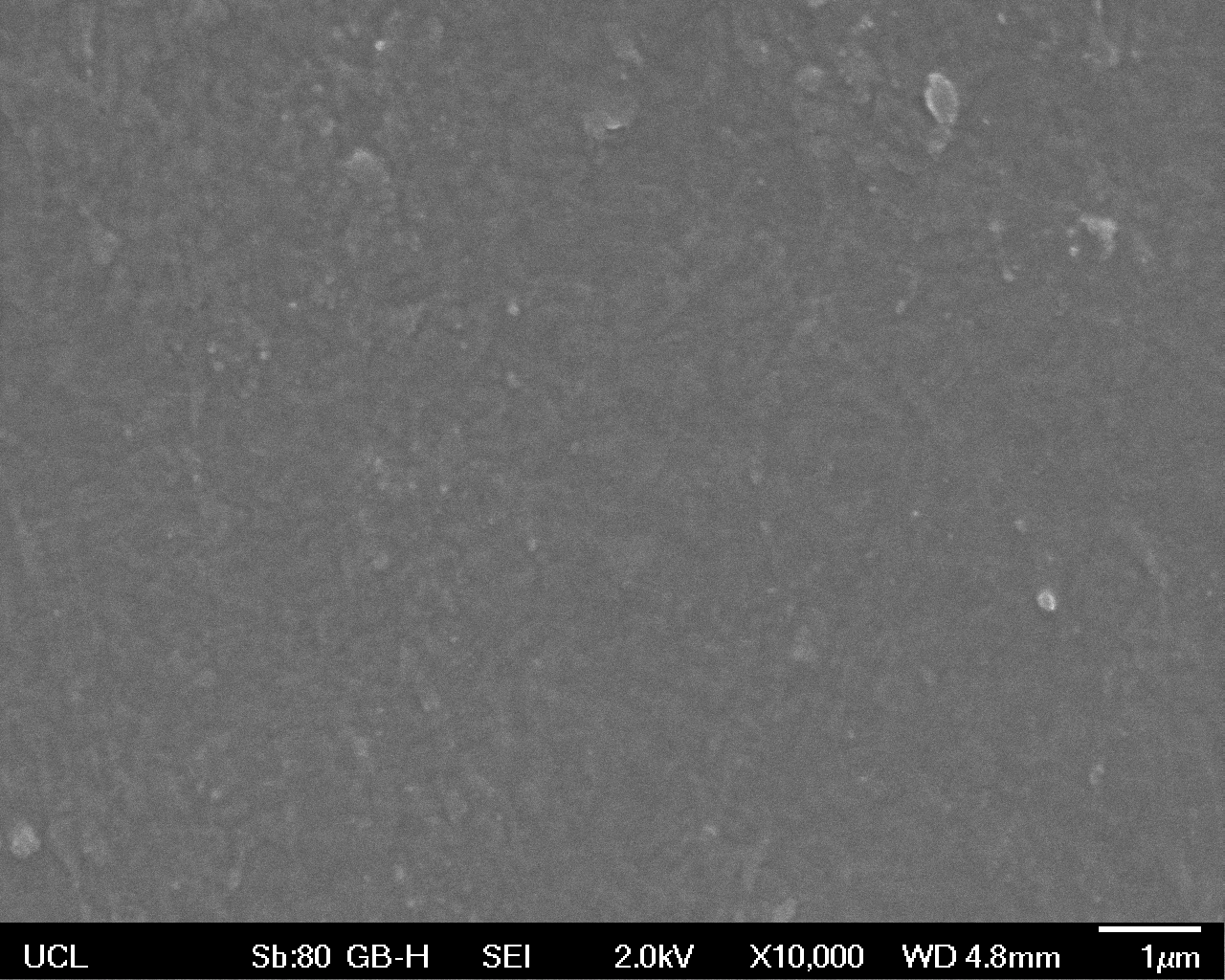 |
 |

|
Modelling
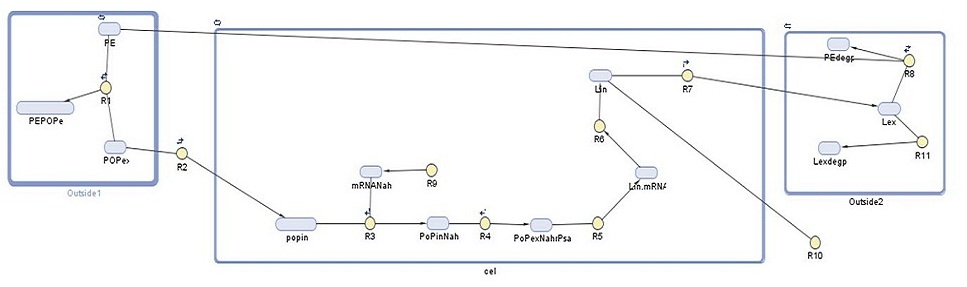
Building upon our earlier cell model for detection of polyethylene, our cell model for the degradation module aimed to show the amount of laccase our bacteria produce and its role in the degradation of one type of microplastic present in the gyre, polyethylene. With this model we aimed to have an idea of how much plastic we could expect our system to degrade. This would then be used in further predictive modelling to calculate the amount of bacteria needed for the construction of Plastic Republic. The following diagram shows the degradation system, with each reaction Ri and each DNA species explained below.
Our cell model was made in SimBiology, which is a MATLAB extension used to run simulations of the reactions in a specified system. It shows the following reactions taking place in three 'compartments', with each DNA species and reaction explained in further detail below:
1. Outside1: in this compartment we see the association, adherence, and dissociation of persistent organic pollutants (POPs) from polyethylene (PE) (R1). We rely on the POPs to induce our degradation system, increasing from 105 to 106 the specificity to our system.
2. Cell: NahR is a constitutively produced mRNA product (R9). When POP diffuses into the cell (R2), it forms a complex with NahR (R3) which then binds to the pSal promoter (R4) to induce production of laccase (R5, R6). Around 20% of this laccase diffuses outside of the cell (R7), the rest degrades (R10).
3. Outside2: in this compartment, the external laccase degrades PE (R9). Some of this external laccase is degraded (R11).
Results
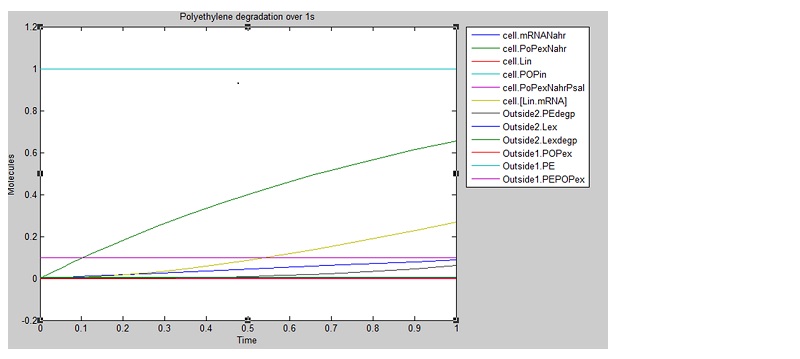
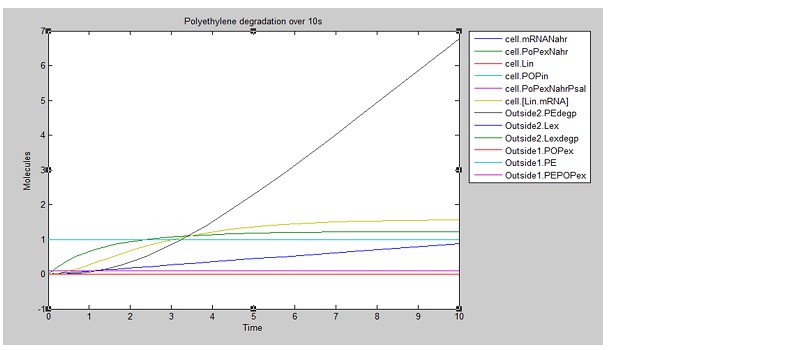
We ran three simulations in SimBiology, each over a different timespan to analyse how laccase production changed over time. In the first second of laccase production, we see polyethylene degradation beginning from around 0.6 sec:
After 10 seconds, the first few molecules have been degraded. The constant diffusion of POPs into the cell is essential for the continuing degradation:
From these results, we can see that our bacteria will degrade polyethylene linearly after an initial non-linear increase in degradation. However, these graphs do not indicate which factors, such as initial concentrations, parameter values and environmental conditions are affecting degradation. For this reason, we decided to run a sensitivity analysis on our model to test the robustness of the model.
Conclusion
Through the use of a simple quantitative assay, it has been ascertained that E. coli, transformed with our construct has a significantly increased extracellular laccase activity when compared with a control cell line. Combined with the results from the scanning electron microscope, which indicate surface degradation of polyethylene in a relatively short timespan, indicates that this BioBrick holds powerful potential for the break down of polyethylene.
| None |