Part:BBa_K510042
pUC18Sfi-miniTn7BB-Gm-Lux
pUC18Sfi-miniTn7BB-Gm-Lux is a vehicle vector for the minitransposon miniTn7BB-Gm-Lux. This transposon is a part of the [http://2011.igem.org/Team:UPO-Sevilla/Foundational_Advances/MiniTn7/Overview miniTn7 BioBrick toolkit], a set of plasmids harboring Tn7 derivatives that can be used for integration of BioBricks in single copy at a conserved target (the attTn7 site), in multiple bacterial genomes.
The mini-Tn7-Gm-Lux artificial transposon has been constructed based on pUC18Sfi-miniTn7-Gm (BBa_K510000) by inserting a pBad/Lux operon (BBa_K325909) in its BioBrick cloning site (BCS). This plasmid can be used for visualization purposes or to brand a strain because of its insertion in the genome of the working organism and split the antibiotic resistance cassette with the temporary expression of Flp recombinase.
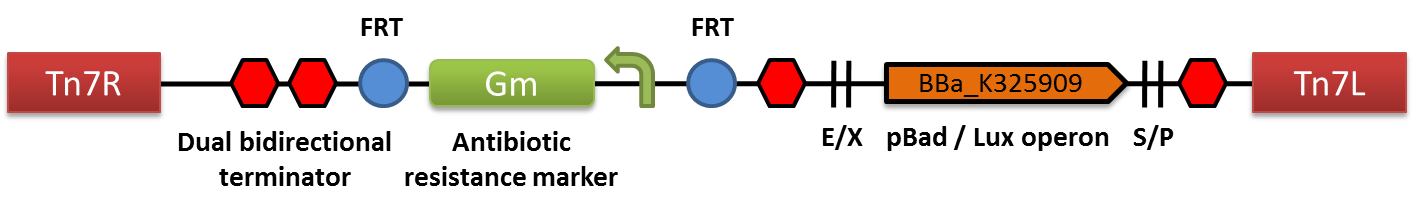
The structure of the mini-Tn7BB-Gm-Lux transposon is shown in the figure below. Tn7R and Tn7L are the right and left ends of Tn7 transposon, respectively, required for recognition of the transposase machinery and transposition. FRT is the Flp recombinase target site for excision of the resistance once the transposon is inserted in the genome of a living organism. The gentamycin resistance cassette (Gm) is flanked by restriction sites in order to facilitate the generation of variants with different antibiotic resistance markers. The pBad/Lux cassette has been inserted in the BioBrick cloning site (BCS).
pUC18Sfi is a pUC18-derived cloning vector harboring an ampicillin resistance marker, a high copy number mutant pMB1 replication origin and the pUC18 multi-cloning site flanked by two SfiI restriction sites. Because of its high copy number and its ability to replicate in any E. coli strain and in other enterobacteria, pUC18Sfi is suitable for maintenance of miniTn7 transposon derivatives, and for simple plasmid preparation and genetic manipulation. pUC18Sfi can be used to deliver transposons by transformation in bacteria in which it does not replicate (i.e., non-enteric bacteria), but not in the enterics.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 4367
Illegal suffix found in sequence at 1 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 4367
Illegal NheI site found at 5601
Illegal NheI site found at 8567
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 4373 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 4367
Illegal BglII site found at 3051
Illegal BglII site found at 3322
Illegal BglII site found at 3608
Illegal BglII site found at 7565
Illegal BamHI site found at 5540
Illegal XhoI site found at 8395 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 4367
Illegal suffix found in sequence at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 4367
Illegal XbaI site found at 4382
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal AgeI site found at 5375 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1681
Illegal BsaI site found at 9954
Illegal BsaI.rc site found at 6963
Illegal SapI site found at 5357
Illegal SapI.rc site found at 2763
Illegal SapI.rc site found at 10279
| None |