Part:BBa_K431007
Alcohol Oxidase 1 promoter
Pichia pastoris initially gained attention as an expression system after recognition of its methylotrophic capabilities and the high levels of alcohol oxidase (AOX) present when induced by methanol. By isolating the promoter (PAOX1)responsible for AOX1 production, this system can be utilized to obtain high yields of heterologous protein. The promoter provides not only high expression but very tight regulation as well. Vectors providing protein expression under the control of PAOX1 are the primary system used for P. pastoris.
ShanghaiFLS_China 2019 characterization
Methanol Induction of the AOX1 Promoter
The promoter of the AOX1 gene (alcohol oxidase 1, gene symbol: PAS_chr4_0821) (PAOX1), homogeneous to Pichia pastoris GS115, is a very strong promoter that is tightly regulated by the carbon source present in the media: it is strongly inhibited by glucose and glycerol, but significantly induced when methanol is present as the only carbon source. This characteristic allows P. pastoris to be a very preferable strain for regulated bioreactor production. It can also be used as an efficient expression system for heterologous proteins.
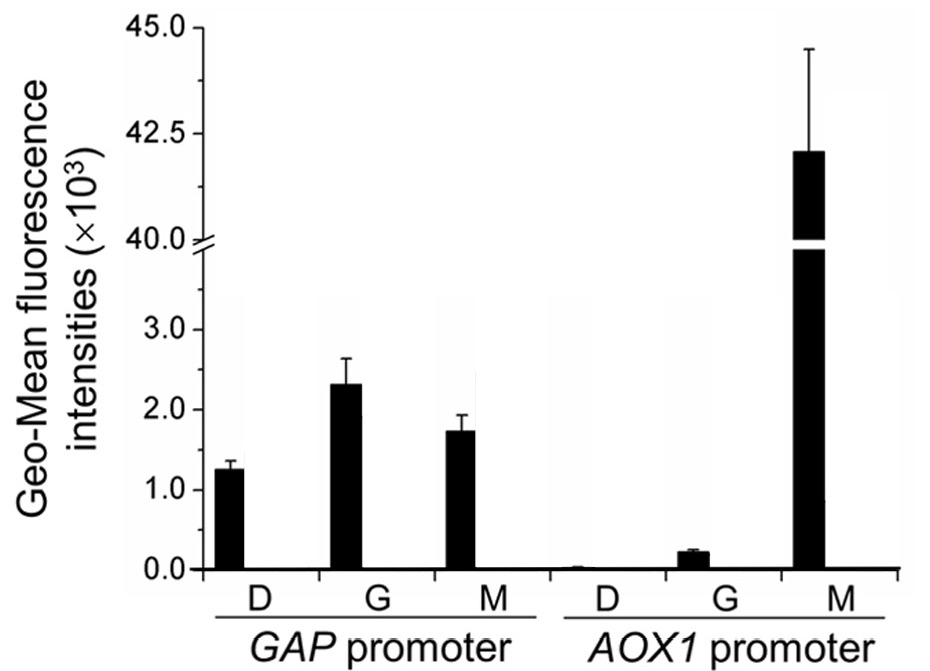
In Wang et al., 2016, the regulation of the PAOX1 is characterized by GFP fluorescence and is compared to that of the GAP promoter (PGAP), a strong constitutive promoter in P. pastoris. Specifically, GFP is expressed by each of the promoters and these constructed strains of P. pastoris are incubated for 12 hours in YNB medium (0.67% yeast nitrogenous base without amino acids) and 50 mg/mL histidine supplemented with 1% glucose (YND), 1% glycerol (YNG), or 0.5% methanol(YNM). The GFP fluorescence intensity is measured by an enzyme-labeled instrument (Synergy 2 by BioTek Instruments) and normalized to unit cell concentrations (in terms of arbitrary units as represented by OD600 measured by the enzyme-labeled instrument). The geometric mean of the normalized data is then calculated and analyzed.
As illustrated by Figure 1, the activity of the pGAP remains largely unchanged regardless of the carbon source in the media. The activity of pAOX1, however, is minimal in glycerol and glucose media, yet very significant in methanol media.
In wild type P. pastoris, alcohol oxidase 1 allows the methylotrophic yeast to metabolize methanol. The regulation of the PAOX1 indicates high specificity in the expression of alcohol oxidase 1 and therefore makes P. pastoris a very attractive candidate for regulated fermentation on an industrial scale, especially if the production of a certain chemical needs to be precisely controlled: the protein expressed by PAOX1 can be relatively simply yet precisely regulated by shifting the carbon source present in the media.
This experiment is published in Wang et al.,2016, and is corroborated by the results from Liang et al., 2012 and Shi et al., 2019.
in trans Regulation of PAOX1
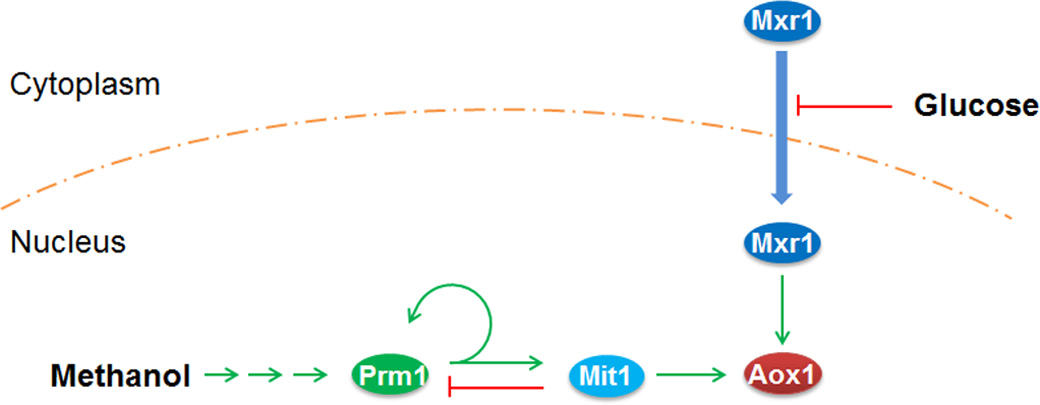
In Wang et al,.2016, it is also experimentally demonstrated how PAOX1 is in trans regulated upon methanol induction. Various knock-out and knock-in strains were produced and the mRNA transcription levels of the proposed transcription factors were evaluated to illustrate the regulation.
Based on the interpretation in this paper, upon methanol induction, transcription factors Mxr1 and Prm1 are activated. The former derepresses PAOX1 while the latter activates PAOX1. The activation of PAOX1 is further amplified via the self-activation of Prm1 (Prm1 activates its own promoter, PPRM1) and the activation of Mit1 by Prm1. Mit1 is a third transcription factor that activates PAOX1. Given that PAOX1 expresses alcohol oxidase 1 in P. pastoris, the key protein that allows it to metabolize methanol, this strong activation allows the methylotrophic yeast to rapidly respond to the change of carbon source from glucose, etc. to methanol. After PAOX1 is strongly activated, over activation is prevented via the suppression of PPRM1 by Mit1. This regulatory pathway is illustrated in Figure 2.
Experimental Data Demonstrating PAOX1 Activity in Methanol Media
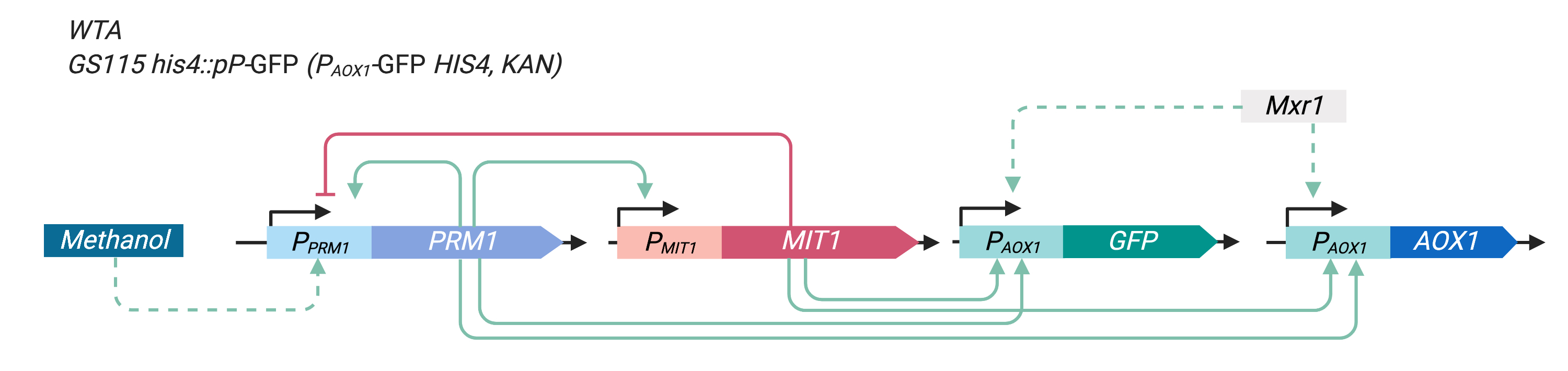
In our experiment, we integrated PAOX1-GFP (yEGFP3 expressed by the PAOX1, BBa_K3239007) into wild type Pichia pastoris GS115 to reflect the activity of PAOX1wild type P. pastoris GS115.
This is valuable given that the expression of AOX1 is critical to the methanol metabolizing ability of P. pastoris, and in certain uses functional proteins such as the insulin precursor expressed by PAOX1 (PAOX1-IP) is integrated into P. pastoris much in the same way as how we integrated PAOX1-GFP into the yeast.
Figure 3 illustrates the modified P. pastoris GS115 construct after PAOX1-GFP integration.
Methods
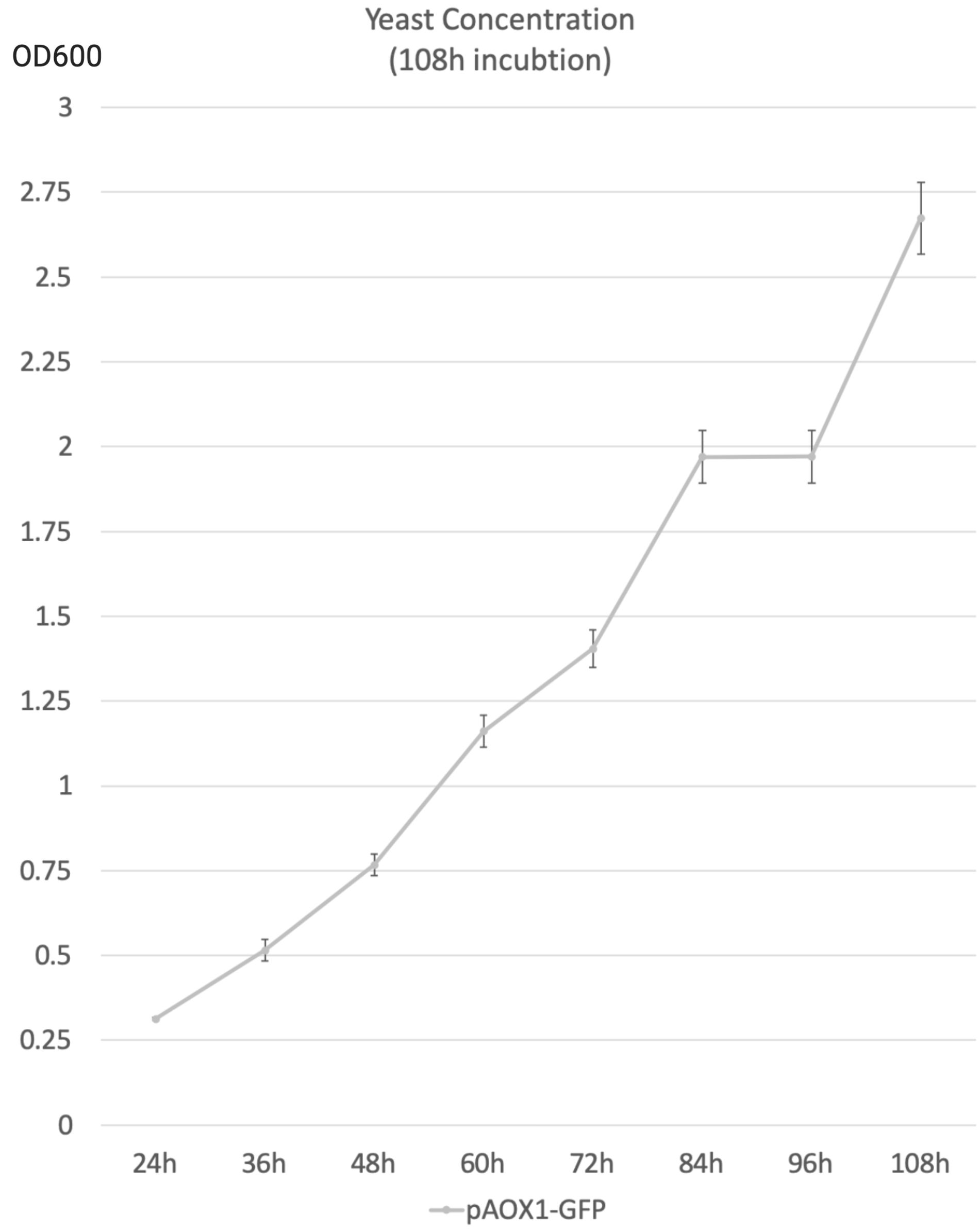
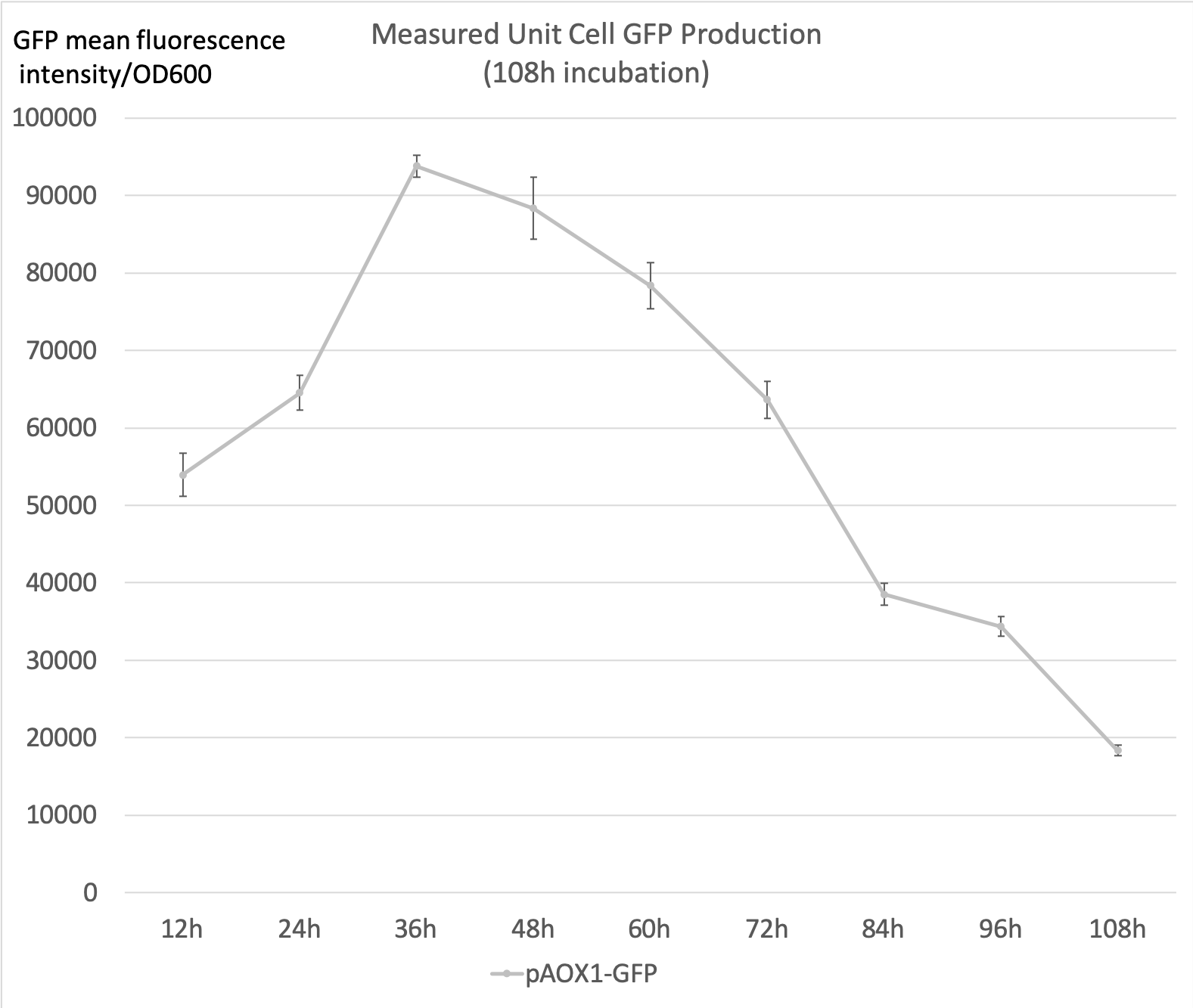
- The following data were acquired after 108h 50mL YNM (1.34% yeast nitrogenous base with 0.5% methanol, 50µg/mL histidine and 40µg/mL biotin) incubation, sampled every 12h. The starting concentration was 1 OD600.
- 0.5% Methanol was added to the media every 24 hours to maintain the methanol concentration.
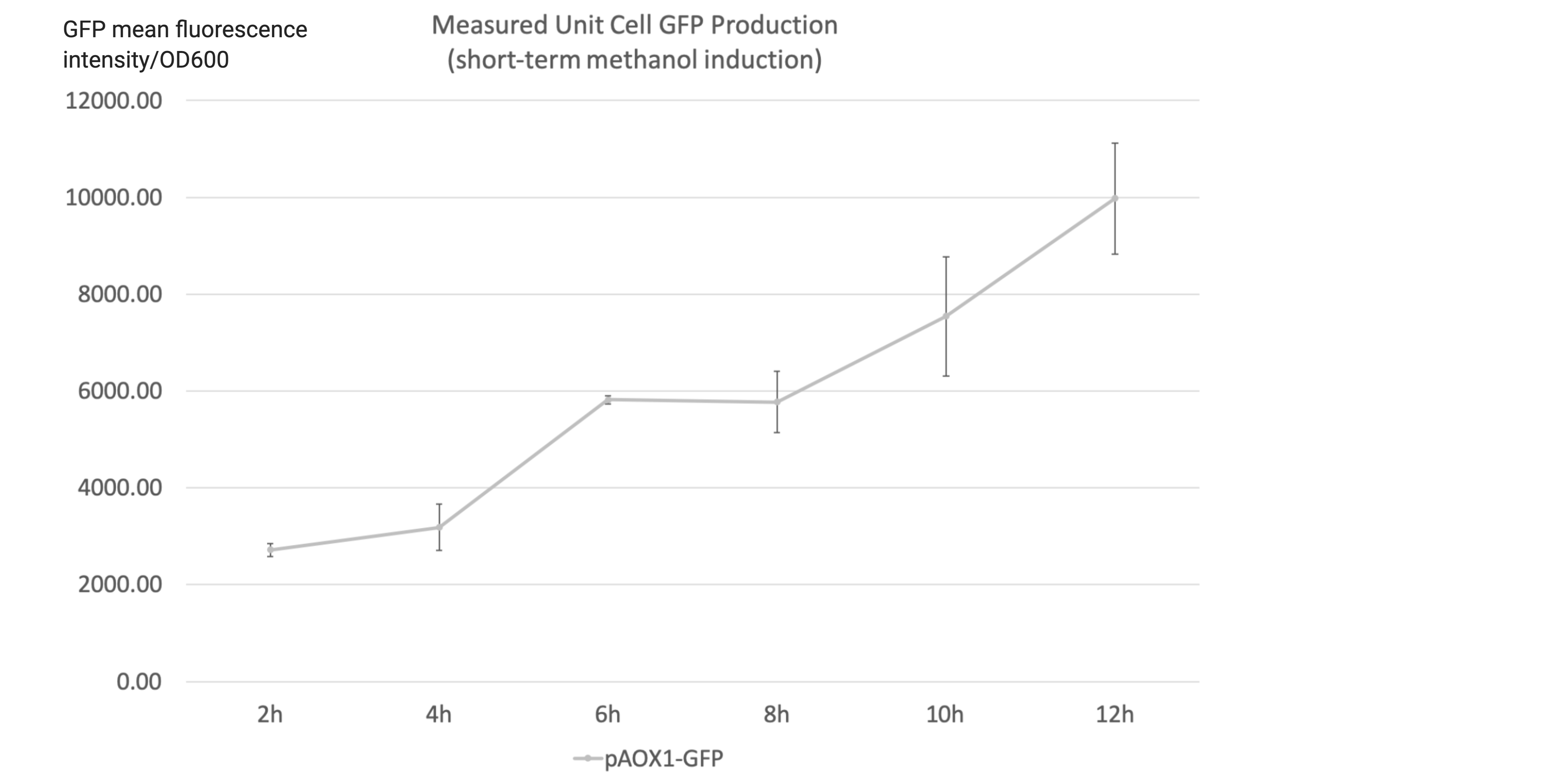
- A short-term methanol induction experiment was performed after the 108h incubation to characterize the instant response to methanol induction after the yeast cells are accustomed to methanol media, hence indicating the overall induction activity of the construct. The yeast cells were collected, rinsed and stored at 4˚C overnight to allow GFP to fully degrade. The strains were then incubated in higher-concentration 50mL YNM media (1.34% yeast nitrogenous base with 0.75% methanol, 50µg/mL histidine and 40µg/mL biotin) for 12 hours and sampled every 2 hours. The starting concentration was 3 OD600.
- Two to three parallels were performed for each strain.
- We used a Biotek Synergy 2 plate reader to measure the GFP mean fluorescence intensities and the OD600 absorbance of our samples.
- Methanol concentration measurements were performed with a Shenzhen Sieman M100 Biosensors Analyzer.
- For teams wishing to compare their data to ours, we've provided our data along with our plate reader calibration data in our wiki. Feel free to give it a check!
Data
- OD600 is used as an indicator of yeast concentration in the experiment.
- Measured unit cell GFP production is the ratio of the GFP mean fluorescence intensities to the 0D600 absorbance measured by the plate reader. It shall serve as an indicator of the expression level of GFP at the sampled time point.
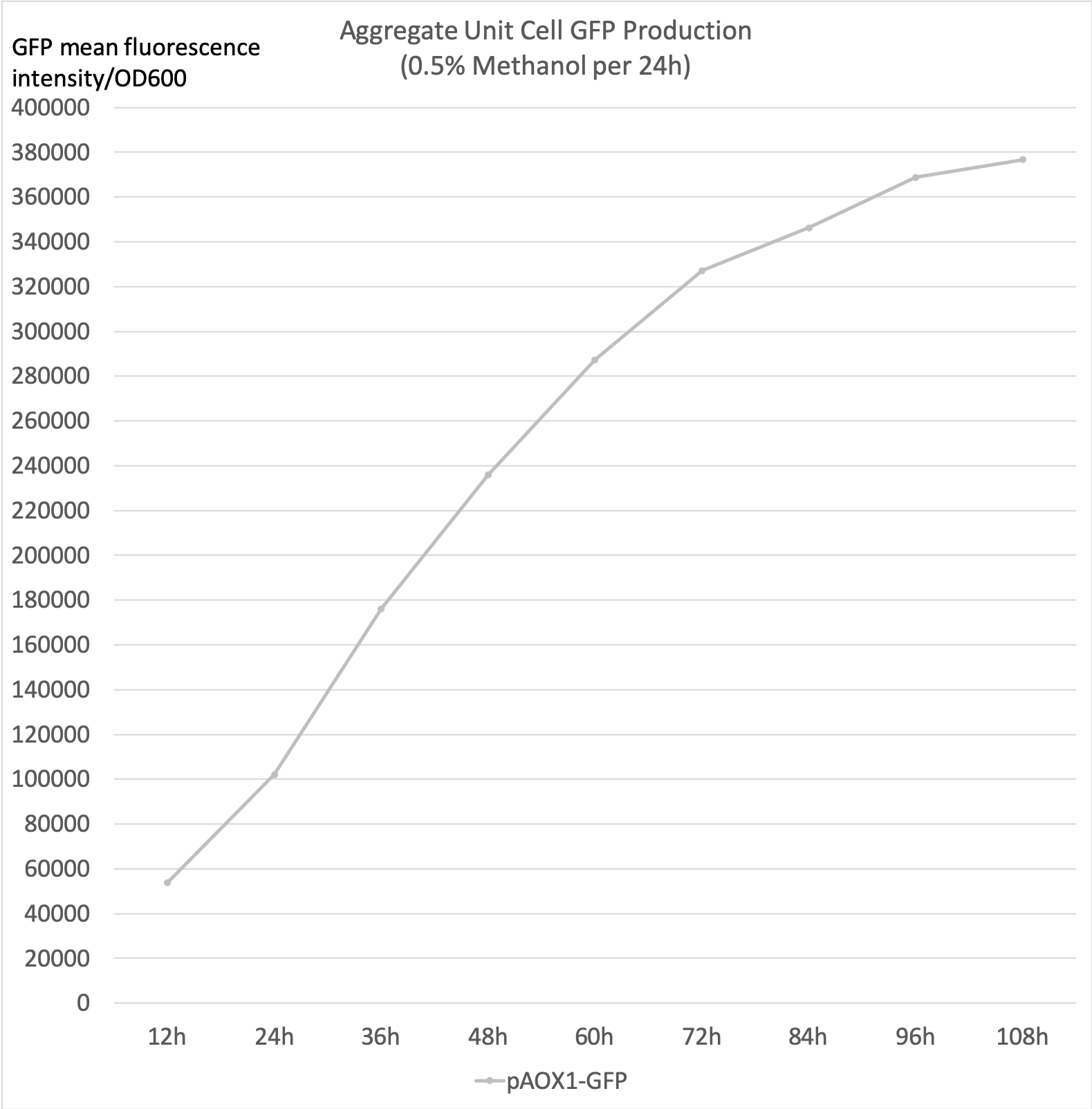
- Aggregate unit cell GFP production is the calculated total unit cell GFP fluorescence intensity. It accounts for the GFP that has degraded over the incubation period to more accurately reflect the aggregate production rate.
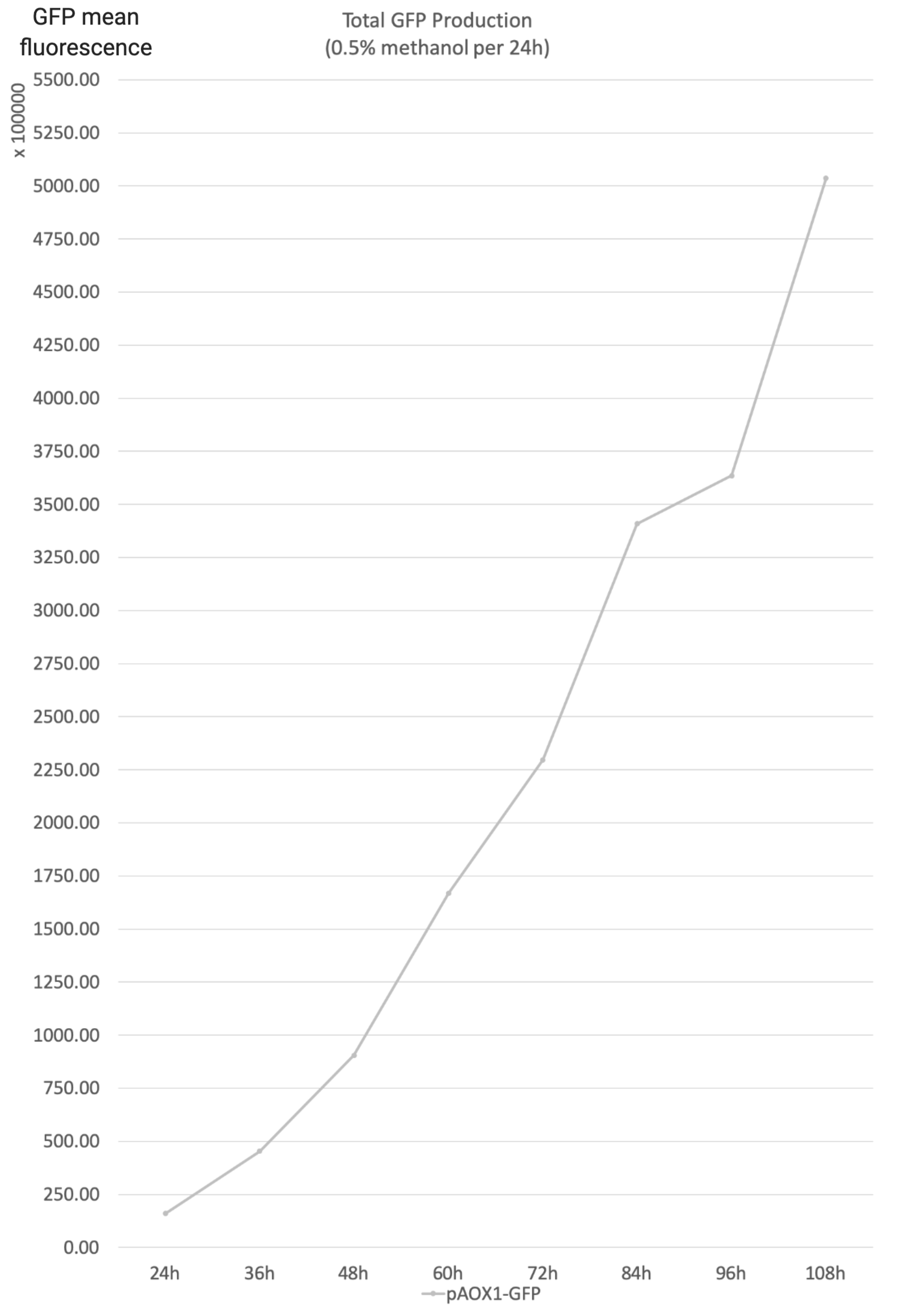
- Total GFP production is the product of the yeast concentration and the aggregate unit cell GFP. It reflects the total GFP production of the reaction system of the strain.
For detailed use of PAOX1 in our experiments, please check our wiki!
https://2019.igem.org/Team:ShanghaiFLS_China
Reference
Wang, X., Wang, Q., Wang, J., Zhou, M., Shi, L., Zhou, X., … Shen, W. (2016). Mit1 Transcription Factor Mediates Methanol Signaling and Regulates the Alcohol Oxidase 1 ( AOX1 ) Promoter in Pichia pastoris. Journal of Biological Chemistry, 291(12), 6245–6261. https://doi.org/10.1074/jbc.m115.692053
Liang, S., Wang, B., Pan, L., Ye, Y., He, M., Han, S., … Lin, Y. (2012). Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics, 13(1). https://doi.org/10.1186/1471-2164-13-738
Shi, L., Wang, J., Wang, X., Zhang, Y., Song, Z., Cai, M., & Zhou, X. (2019). Transcriptome and metabolome analyses reveal global behaviour of a genetically engineered methanol-independent Pichia pastoris strain. Process Biochemistry, 76(August 2018), 46–54. https://doi.org/10.1016/j.procbio.2018.10.014
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//promoter
| None |