Part:BBa_K4292018
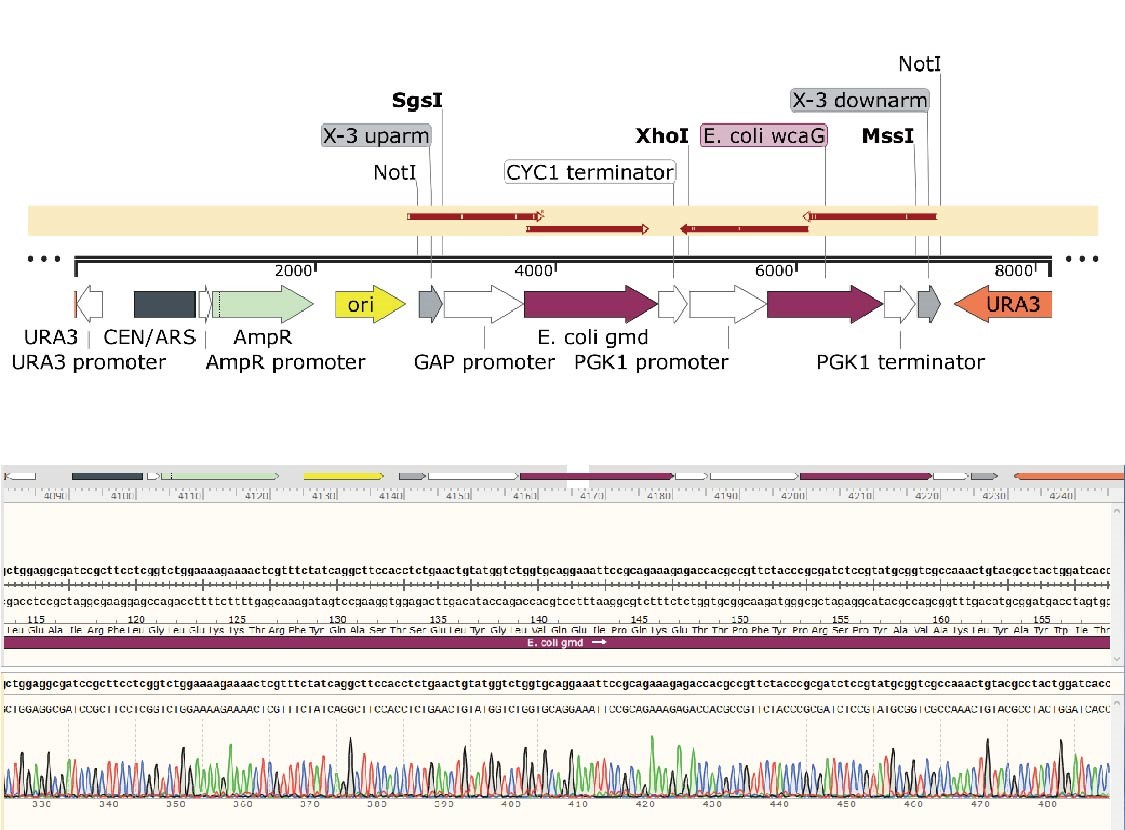
X-3 uparm-GAP promoter-E. coli gmd-CYC1 terminator-PGK1 promoter-E. coli wcaG-PGK1 terminator-X-3 do
X-3 uparm-GAP promoter-E. coli gmd-CYC1 terminator-PGK1 promoter-E. coli wcaG-PGK1 terminator-X-3 dowmarm
Contribution
Breast milk oligosaccharides, also known as human milk oligosaccharides (HMOs), are the third abundant solid component in breast milk after lactose and fat. Over 200 sugar compounds were identified in HMOs, among which 2′-Fucosyllactose (2’-FL) has the highest proportion in HMOs (more than 30%). Studies have proved that 2'-FL not only promotes the development of immune system and a gut microbiota, but also helps to prevent allergic disease and promotes brain function and cognitive development. Moreover, 2'-FL can be used as an additive in the infant formula which is closer to breast milk in nutrients. Thus, producing the high yield of 2'-FL draws more and more attention.Breast milk oligosaccharides, also known as human milk oligosaccharides (HMOs), are the third abundant solid component in breast milk after lactose and fat. Over 200 sugar compounds were identified in HMOs, among which 2′-Fucosyllactose (2’-FL) has the highest proportion in HMOs (more than 30%). Studies have proved that 2'-FL not only promotes the development of immune system and a gut microbiota, but also helps to prevent allergic disease and promotes brain function and cognitive development. Moreover, 2'-FL can be used as an additive in the infant formula which is closer to breast milk in nutrients. Thus, producing the high yield of 2'-FL draws more and more attention.
At present, there are three methods for synthesizing 2'-FL, including chemical synthesis, enzymatic synthesis, and whole-cell biosynthesis. However, there are some limiting factors for the chemical synthesis and enzymatic synthesis including the toxic reagents and cost of precursor. For instance, using the enzyme-catalyzed synthesis route, the yield of enzyme-catalyzed synthesis of 2'-FL is not high due to the expensive substrate guanosine 5’-diphosphate-L-fucose (GDP-L-fucose). The substrate GDP-fucose is extracted from cellular which is not suitable to reuse it for large-scale production. In contrast, the whole-cell biosynthesis method is relatively low-cost and mainly performed in engineered E.coli to produce 2'-FL. But, for large-scale fermentation, there is possible that endotoxin contamination. For sake of safety, we use the S. cerevisiae to produce 2'-FL. In addition, sweet potato residues contain abundant glucose with the relative content of 56% and can be used as substrate for large-scale fermentation.
Thus, in this project, we designed and synthesized an engineered S. cerevisiae cellular factory producing 2'-FL from sweet potato residues by fermentation. On one hand, producing 2'-FL in S. cerevisiae is a relatively safe cellular factory to allow the large-scale fermentation. On the other hand, it is promising that using sweet potato residues to further extend industrial chain, improve the utilization value of inferior biomass resources and cut the cost. Thus, in this project, we designed and synthesized an engineered S. cerevisiae cellular factory producing 2'-FL from sweet potato residues by fermentation. On one hand, producing 2'-FL in S. cerevisiae is a relatively safe cellular factory to allow the large-scale fermentation. On the other hand, it is promising that using sweet potato residues to further extend industrial chain, improve the utilization value of inferior biomass resources and cut the cost.
Our team aims to produce 2'-FL in S. cerevisiae for providing the economic strategy. In terms of raw material, there are two pathways to produce GDP-L-fucose from GDP-D-mannose (called de novo pathway) or L-fucose (called salvage pathway). In the de novo pathway, the GDP-D-mannose converts 2'-FL and secreted into medium involving three enzymes gmd, wcaG, and wbgL, as well as transporter lac12.
In order to obtain the high yield of 2'-FL, we introduced the four exogenous gene including Lac12, gmd, wcaG, and Wbgl into S. cerevisiae genome using CRISPER-cas9. As shown in Figure1, expression of Lac12 (lactose permease) from Kluyveromyces lactis is a transporter to facilitate lactose transporting out of the cytoplasm. Other three genes, gmd, wcaG and Wbgl, are encoding the important enzymes involving the 2'-FL synthesis. Amongst these enzymes, gmd and wcaG are converted the abundant intracellular GDP-mannose in S. cerevisiae into GDP-fucose. Further, 2'-FL can be produced from GDP-fucose by expression of Wbgl and metabolization. In this work, we used sweet potato residues as main carbon source.
Engineering Success
pHCas9-Nours plasmid serves as a DNA template and amplified PCR to obtain the XI-2 and X-3 site target gene integration plasmid. The gmd, lac12, wbgl and wcaG gene fragments were amplified by PCR using the X-3-gmd-wcaG plasmid and the XI-2-wbgL-lac12 plasmid as template.
The DNA fragments gmd-wcaG and wbgL-lac12, as well as XI-2 and X-3 integration plasmids were digested with NotI and XhoI to form the cohesive ends, respectively. Then, the DNA fragments and vector were joined together by the ligase. The recombinant plasmid transformed into the competent cells and verified by colony PCR and Sanger sequencing. After that, the corrected gmd-wcaG and wbgL-lac12 DNA fragments, which were digested with NotI, and gRNA were introduced into the yeast that already contains the Cas9 expression plasmid using lithium acetate transformation method. These colonies were used colony PCR to verify whether the colony’s genome carried recombinant DNA fragments, and the positive transformants further confirmed through Sanger sequencing.
Finally, the positive colony incubated in the shaker, enabling produce 2'-FL in YPD30L2 medium and sweet potato mash. Finally, we could obtain high yield of 2'-FL in the engineered yeast.
Gene integration plasmid construction: Wbgl and lac12; gmd and wacG
We constructed two plasmids including four genes WbgL, lac12, gmd, and wacG which are key genes to produce 2'-Fucosyllactose(2'-FL) in a yeast cellular factory. In addition, we need to added the promoter and terminator to flanking regions of these exogenous genes in order to facilitate expression in the engineered yeast. The components were incorporated into the integration backbone plasmid with NotI and XhoI sites. Firstly, we constructed the integration backbone plasmids XI-2 and X-3. These colonies were verified by colony PCR and Sanger sequencing and the results were shown as follows.
A.Validation of the construction of XI-2 site integration backbone plasmid
XI-2 site target gene integration backbone plasmid, containing 200 bp upstream and downstream homology arms of chromosome XI-2 site. Randomly pick 10 transformants from the LB-Amp plate, and use the upstream and downstream primer pairs of the homology arm to verify whether the fragment inserted. The target band is about 450 bp. It’s possible that 2, 3, 4, 5, 9 and 10 transformants has inserted the DNA fragments as shown in Figure 4.
The plasmids of transformants 2, 3, 4, 5, 9, and 10 were extracted and further verified by NotI digestion (Figure 5). If the cut band size is correct as shown in Figure 5, we picked No. 10 to perform Sanger sequencing.
The sequence of the transformant plasmid No. 10 is no mutation and mismatch as shown in Figure 6 indicating we successfully constructed XI-2 site integration backbone plasmid.
B. Validation of the construction of X-3 site integration backbone plasmid
The X-3 site contains 200 bp of homology arms, which on the upper and lower sides of the chromosome X-3 site. 10 transformants were randomly picked from the LB-Amp plate, and the upstream and downstream primer pairs of the homology arms were used to verify whether the fragment inserted. The target band was about 450 bp. As shown in Figure 7, except for 6 and 7, the rest of the transformants might insert fragments.
The plasmid of transformants 2, 3, 5, and 8 were extracted and further verified by by Not1 digestion (Figure 8). The target band was 3762+430 bp. Thus, we picked No. 5 for sequencing, and the results are shown in Figure 9.
The sequence alignment well matched, indicating that the X-3 site integrated backbone plasmid was constructed successfully. Secondly, the wbgL-lac12 and gmd-wcaG incorporated into the integration plasmids XI-2 and X-3, respectively. The verification results were shown as follows.
C. Validation of the construction of XI-2-wbgL-lac12 plasmid
In the XI-2-wbgL-lac12 plasmid, the wbgL and lac12 gene expression cassettes are inserted between the XI-2 homology arm, in different orientations. The wbgL+lac12 gene expression plasmid for XI-2 site integration was constructed by a two-step digestion cloning method. After wbgL gene expression cassette was integrated, lac12 inserted though Xho1 and BamH1 digestion sites.
Plasmids of 10 transformants were extracted and verified by Xho1+BamH1 double-enzyme digestion (Figure 10). The positive transformant band was 6007+2635 bp, and the correct NO.11 was selected for sequencing. The results were shown as Figure 11.
The sequence alignment results showed well matched, indicating that the XI-2-wbgL-lac12 plasmid was constructed successfully.
D. Validation of the construction of X-3-gmd-wcaG plasmid
In the X-3-gmd-wcaG plasmid, the gmd and wcaG gene expression cassettes are inserted between the upstream and downstream of the X-3 homology arm, in the same direction. The gmd+wcaG gene for X-3 site integration was constructed by a two-step digestion cloning method. After the gmd gene expression cassette was integrated, the wcaG was introduced using Xho1 digestion site.
Plasmids of 12 transformants were extracted and verified by Xho1+Mss1 double-enzyme digestion (Figure 12). The positive transformant band was 6220+1896 bp, and the No.11 plasmid with correct digestion was randomly selected and sent for sequencing.
The details of sequence alignment showed there is no mutation, indicated that the X-3-gmd-wcaG plasmid was constructed successfully.
Improvement of an Existing Part
Using CRISPR Cas-9 technology to integrate the target genes into the genome of S. cerevisiae
The constructed X-3-gmd-wcaG and XI-2-wbgL-lac12 plasmids were digested with Not1, respectively, and the large fragments were extracted from the gel. Using the lithium acetate transformation method, together with the gRNA, they were introduced into the yeast competent cell that already contains the Cas9 expression plasmid. In yeast CCTCC M94055, colony PCR was used to verify the integration of exogenous genes.

Colony PCR was used to verify whether the X-3 and XI-2 loci gene fragments were integrated into S. cerevisiae strains. The results are shown in Figure 14. The integrated copy number of the four transformants was verified using different primer pairs. If the internal primers (inner-primer pairs) can amplify the target band, and the outer primers (outer-primer pairs) cannot amplify the target band with the size of the integration homology arm, it means that 2 copies have been integrated. If the primers outside the site amplify the target band of the size of the integration homology arm, it means that 1 copy of the target gene has been integrated. Based on the above analysis, we judged that the middle transformants 6 and 7 have clearly integrated one copy of the target gene, and can be tested for subsequent experiments.
The engineered strain utilizes synthetic medium to produce 2'-FL
The productivity of 2'-FL by recombinant yeast in the synthetic medium YPD30L2 was tested. As shown in Figure 15, 30 g/L glucose was completely consumed within the initial 4 h, and then the strain began to use the produced ethanol as a carbon source, showing a secondary growth state (Fig. 12A). The initial addition of 2 g/L of lactose was undetectable after 48 h, resulting in about 0.7 g/L of 2'-FL (Fig. 12B). The theoretical conversion rate of lactose to 2'-FL was 100%. Part of 2'-FL accumulated intracellularly and failed to be effluxed into the medium.
Production of 2'-FL from sweet potato residues by engineered strains
The productivety of 2'-FL by recombinant yeast from sweet potato residues was tested, as shown in Figure 16. Generally, the growth of the strain was slightly worse than that of the synthetic medium (Figure 16A), which may be due to the presence of some inhibitor for yeast growth in the sweet potato residues. The presence of lactose was not detected at 48 h, and the final yield was about 0.6 g/L 2'-FL (Figure 16B).
By retrieving the igem library, we found the data numbered BBa_K4085015, by Group: iGEM21_Ulink-SIP (2021-10-21) Uploaded, our experimental results are better than the experimental data already uploaded in the igem library, which also proves the advantages of our experimental design scheme. We hope that our experiments can provide reference for other igem teams and provide guidance for subsequent industrial production improvements.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 889
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1146
Illegal BsaI.rc site found at 1306
| None |