Part:BBa_K4248005
LL-37-Linker-Sparamosin26-54
LL-37-Linker-Sparamosin26-54
Contribution
We are the Pepsick iGEM team dedicated to developing antimicrobial peptide products for cleaning fish tanks. In order to carry forward the spirit of iGEM, and inherit and spread the value of iGEM, we specially searched the iGEM Biological Parts library for related antimicrobial peptides and picked BBa_K1162002, Spheniscin-2 antimicrobial peptide from the king penguin (Aptenodytes patagonicus). This is a biological part submitted by iGEM13_Utah_State in 2013, with only DNA sequence information and simple text description information. Our team carried out a comprehensive characterization of this part in the laboratory, adding data from antibacterial testing to dedicate its function and properties in inhibiting bacterial growth. This information can be a good reference for future iGEM teams working on antimicrobial peptides.
What's more, we have added a new fusion antimicrobial peptide, which is formed by fusing LL-37 and Sparamosin26-54 through a protein linker, which can effectively combine the antibacterial functions of LL-37 and Sparamosin26-54, not only inhibiting bacterial growth but also inhibiting fungal growth, that is an original and new form of fusion antimicrobial peptides, which also provides more ideas for future iGEM teams to optimize antimicrobial peptide products.
Engineering Success
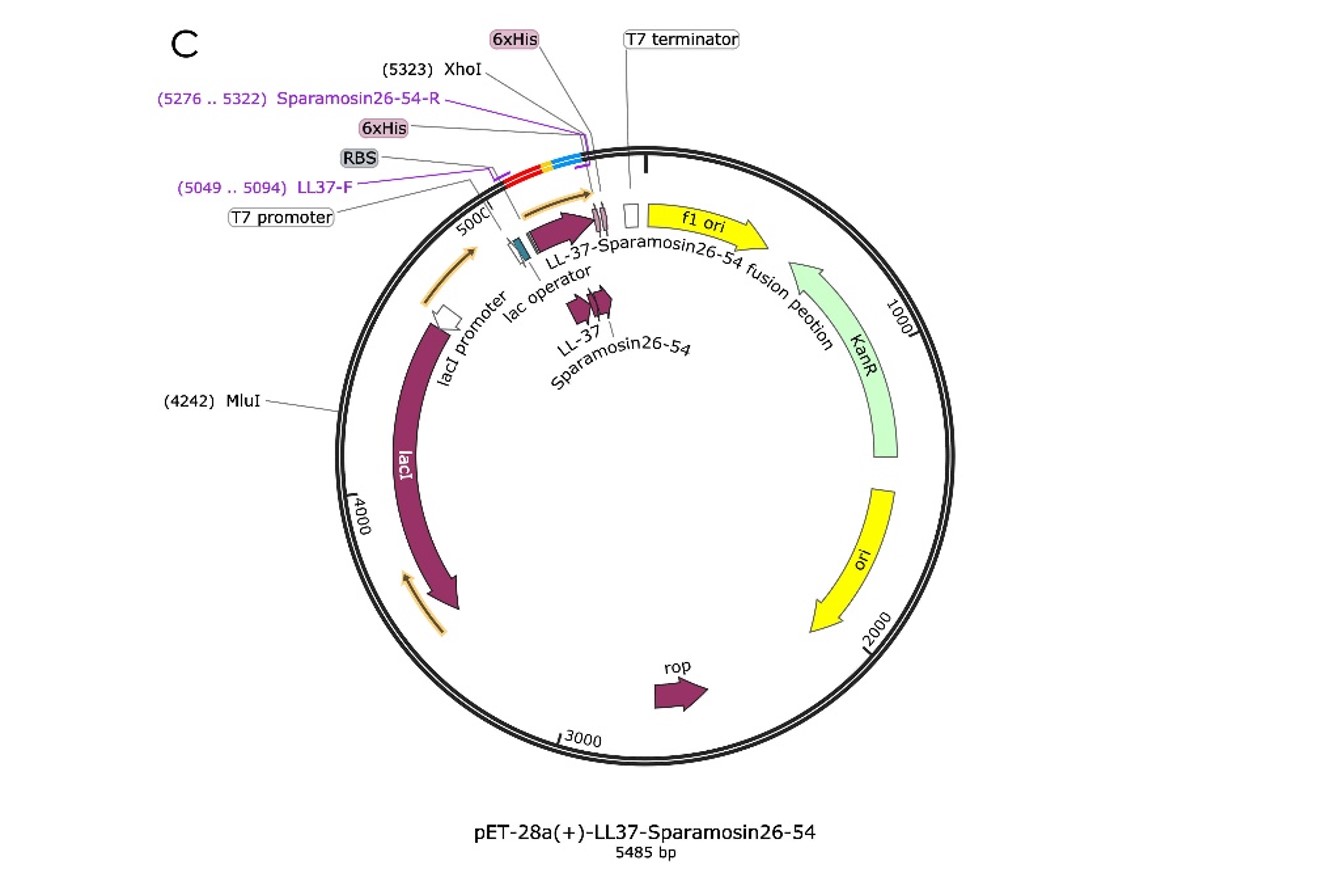
Construction of expression plasmids
We design the plasmid: a fusion AMP that connects LL-37 and Sparamosin26-54 through a linker, which was also inserted into the pET-28a(+) vector for protein expression, as shown in Figure 1.
To build the plasmid, we let the synthetic company synthesize the DNA fragment of Fusion and integrate it into the pET-28a(+) vector. The returned sequencing comparison results showed that there were no mutations in the ORF region (Figure 2.). Thus, the expression plasmids were successfully constructed. And the last step was extracting the recombinant plasmids from E.coli DH5α and transferring them into E.coli BL21(DE3) competent, so that can be used to express AMP proteins.
Protein expression and purification
In order to obtain the AMPs’ proteins, we expanded the culture in the LB medium and added IPTG to induce protein expression when the OD600 reached 0.4. After overnight induction and culture, we collected the cells and ultrasonic fragmentation of cells to release the intracellular proteins. Next, we used nickel column purification to purify the antibacterial peptide protein we wanted. The concentration of protein was measured as: 0.431mg/mL Fusion. At this point, we got the AMPs’ protein solutions we wanted.
Functional test=
To confirm the function of the AMPs to inhibit bacterial growth, we used the real mixed bacteria acquired from Haichang Ocean Park as bacteria, and antibiotics as a positive control for bacteriostatic test experiments.To better show the relationship between the concentration of antimicrobial peptides and the inhibition of bacterial growth, we added 100 μL of mixed bacteria and 100 μL of different concentrations of the AMPs to each tube and repeated them three times for each concentration to form the average data graph with error bars.
As we have seen in Figure 4, the Fusion peptide’s performance (Figure 4C) at each concentration was almost always the best of the three. Furthermore, Fusion widens the gap when the concentration reaches 15 μm. The Fusion peptide did nearly twice as efficient as the rest of the two peptides (Figure 4A, 4B) in the concentration of 15μm, 20μm, and 25μm, proving the success of using the Fusion peptide to enhance our performance of the AMPs.
Improvement of an Existing Part
Our composite part BBa_K4248005 was improved based on the existing part BBa_K1162006. There is a mini-review of the development of LL-37 protein-related parts. In 2009, group iGEM09_Slovenia firstly designed a basic part BBa_K245114, LL37. In 2012, group iGEM12_Trieste designed the part LL37 (BBa_K875009) using Assembly Standard #10 for both the prefix and suffix and optimizing the coding sequence for E.coli. In 2013, group iGEM13_Utah_State further improved the part LL37 (BBa_K1162006) using Assembly Standard #23 and by removing the stop codon at the end of the coding region. In 2019, group Jilin_China 2019 characterized this part to kill C. Albicans. In 2021, group HZAU-China 2021 characterized this part to kill Salmonella Typhimurium SL1344. Today, our team further improved the LL-37 by adding fusing a new peptide Sparamosin26-54 with a Linker between the two APMs. Furthermore, the codon has been optimized for E. coli to develop our new composite part BBa_K4248005.
In order to prove the function of our new fusion part LL37-Linker- Sparamosin26-54 , we expressed and purified the Fusion AMP, LL-37 AMP, and Sparamosin26-54 AMP, then detected the inhibition in mixed bacteria acquired from Haichang Ocean Park. As the result shown above, our Fusion AMP has achieved nearly twice bacteriostatic ability as efficiency as that of LL-37 peptide. Besides, our project aimed to clean fish tanks. And Fusion AMP could well inhibit mixed bacterial growth in fish tanks, which is in accord with our initial expectation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |