Part:BBa_K4130015
Contents
Sar11-5 Aptamer
ssDNA Aptamer for free sarcosine with a 3’ amine modification
This part selectively detects sarcosine with a dissociation constant of 134.8 ± 1.12nM and a literature detection range of 55nM-2uM [10]. Sarcosine (N-methylglycine) is an amino acid derivative that is a byproduct of creatine hydrolysis [1] as well as an intermediate of choline metabolism [2]. In maple trees, it is found to increase in concentration from 0.01uM to 0.12uM [8] as the tree matures and nears new-leaf production (aka “bud break”), containing methyl groups that contribute an off-taste to maple syrup when boiled down from sap [3] Besides maple trees, sarcosine is also a human metabolite, thought to be a marker for prostate cancer [1], and has been investigated for use in treatment against schizophrenia.
Usage and Biology
Aptamers are ssDNA or ssRNA molecules that have selective binding capabilities to target molecules such as proteins or small molecules. These oligonucleotides can be synthesized through PCR and selected in vitro through SELEX [4] Their antibody-like function can be utilized to detect target molecules, particularly in diagnostic biosensor applications known as “aptasensors” [4]. However, aptamers are often easier to synthesize and therefore cheaper than antibodies, in addition to being optimal for specific targeting of smaller molecules [5] such as sarcosine. In context, this aptamer’s function was utilized in an electrochemical aptasensor mechanism, in which the aptamer was pipetted, also known as dropcasting, onto an electrode, followed by an application of current.
Aptamer Specifications
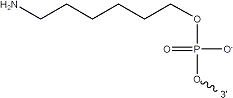
The initial aptamer sequence and structure for Sar09-3 (Fig. 2) was selected based on a previous study that demonstrated effective binding of sarcosine to the picomolar levels [10] After selecting the sequence, in the interest of time and effort, the ssDNA aptamer was ordered to be synthesized by IDT. In addition, the 3’ end of the aptamer sequence was ordered to be modified with an amine group (Fig. 1). This was done to ensure that the aptamers could covalently bond to functional groups present on the surface-deposited nanomaterials applied to screen printed electrodes (SPEs) to improve their performance. Strong electrode-to-aptamer binding, known as immobilization[9], is critical for establishing the accuracy and sensitivity of the aptasensor, since aptamer-to-target binding events incur folding that impedes the current of the electrode, and therefore triggers a signal in the form of a resistance readout.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 67
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 67
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 67
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 67
- 1000COMPATIBLE WITH RFC[1000]
References
“Sarcosine.” Sarcosine - an Overview | ScienceDirect Topics, https://www.sciencedirect.com/topics/chemistry/sarcosine.
“Sarcosine.” National Center for Biotechnology Information. PubChem Compound Database, U.S. National Library of Medicine, https://pubchem.ncbi.nlm.nih.gov/compound/Sarcosine.
Garcia, E Jose, et al. “Metabolomics Reveals Chemical Changes in Acer Saccharum SAP over a Maple Syrup Production Season.” PloS One, Public Library of Science, 20 Aug. 2020, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7444596/.
Liu, Ling Sum, et al. “Recent Developments in Aptasensors for Diagnostic Applications.” ACS Applied Materials & Interfaces, vol. 13, no. 8, 2020, pp. 9329–9358., https://doi.org/10.1021/acsami.0c14788.
Drabek, Rafal. “Aptamers vs. Antibodies - Antibody Alternatives.” Base Pair Biotechnologies, 19 Jan. 2020, https://www.basepairbio.com/aptamers-vs-antibodies/.
Platt, Mark, et al. “Analysis of Aptamer Sequence Activity Relationships.” OUP Academic, Oxford University Press, 12 Nov. 2008, https://doi.org/10.1039/b814892a.
“Attachment Chemistry / Linkers Modifications.” IDT, https://www.idtdna.com/site/Catalog/Modifications/Category/2.
N’guyen, Guillaume Quang, et al. “A Systems Biology Approach to Explore the Impact of Maple Tree Dormancy Release on Sap Variation and Maple Syrup Quality.” Nature News, Nature Publishing Group, 2 Oct. 2018, https://www.nature.com/articles/s41598-018-32940-y.
Oberhaus, Franziska V, et al. “Immobilization Techniques for Aptamers on Gold Electrodes for the Electrochemical Detection of Proteins: A Review.” Biosensors, MDPI, 28 Apr. 2020, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7277302/.
Luo, Yu, et al. “In Vitro Selection of DNA Aptamers for the Development of Fluorescent Aptasensor for Sarcosine Detection.” Sensors and Actuators B: Chemical, Elsevier, 23 Aug. 2018, https://www.sciencedirect.com/science/article/pii/S0925400518315430
| None |