Part:BBa_K3898163
RIDD-MHETase
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 47
Illegal PstI site found at 968 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 968
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1308
Illegal XhoI site found at 1958 - 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 47
Illegal PstI site found at 968 - 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 47
Illegal PstI site found at 968
Illegal NgoMIV site found at 193
Illegal AgeI site found at 1006 - 1000COMPATIBLE WITH RFC[1000]
RIDD-MHETase consists of two components, RIDD (BBa_K3898001) and MHETase (BBa_K3898006). This is a part of our PET degradation three-enzyme complex . Its function is to degree MHET, and produce TPA and EG. Thus MHET won’t come in to PETase’s active pocket as a inhibitor.
SDS-PAGE
The results showed that corresponding target proteins were present in the fragments of the three bacteria. However, due to its smaller molecular weight (11 kDA), RIAD-hydrophobin 4 was not detectable on 10% separation gel.
Channel 1: No expected bands were found in the medium of E.coli BL21 without induced PETase expression after resuspension and centrifugation; Channel 2: No expected bands were found in the precipitate of E.coli BL21 expressing PETase without IPTG induction after centrifugation; Channel 4: M: marker; Channel 3: There were expected bands in the medium of E.coli BL21 where RIDD-PETase expression was induced following by cell disruption and resuspension, but the concentration was low; Channel 4: There were significant correlation bands in the precipitate of induced RIDD-PETase expressing E.coli BL21 after cell disruption, resuspension and centrifugation, exhibiting high concentration. Channel 5: There were expected bands in the medium of E.coli BL21 where RIDD-MHETase expression was induced following by cell disruption and resuspension, but the concentration was low; Channel 6: There were significant correlation bands in the precipitate of induced RIDD-MHETase expressing E.coli BL21 after cell disruption, resuspension and centrifugation, exhibiting high concentration.
Enzyme activity
As is seen in the SDS-PAGE results, after cytoclasis, RIAD-hydrophobin 4, RIDD-PETase, RIDD-MHETase all primarily existed in the precipitate, while almost none were found in the lysis medium. It was known that most proteins in the precipitate are in the form of inclusion bodies and not active. It is difficult to tell whether the target proteins were present in the medium after the cell disruption by direct observation. Therefore, we performed western-blot analysis. The results showed that there was still a small amount of RIDD-PETase in the medium, indicating that instead of waiting for the results of renaturation experiment , the medium can be used to determine the enzyme activity of RIDD-PETase, RIDD-MHETase as crude enzyme solution.
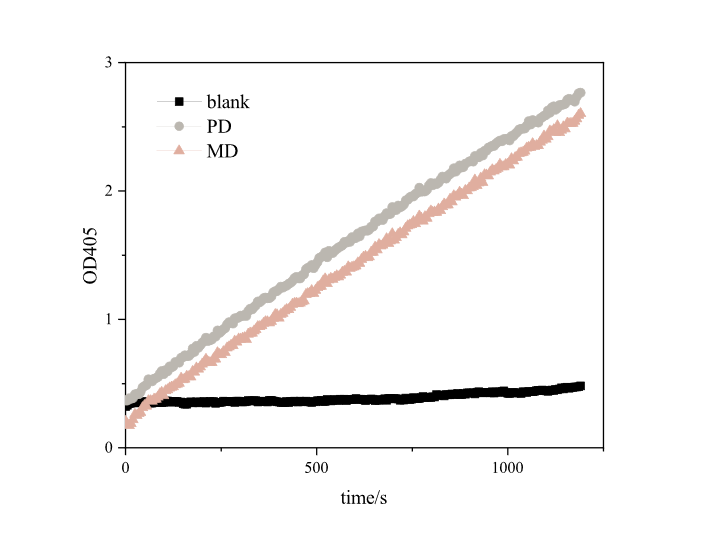
Since it takes a long time to degrade PET plastics, it is impossible to effectively and scientifically measure the enzyme activity of MHETase alone, so we referred to the method of iGEM16_Harvard_BioDesign team and designed a set of methods suitable for our project. The enzyme activity was measured using pNPB, a universal substrate for esterases. Under the reaction of pNPB esterase, p-nitrophenol can be generated, which has a strong absorption peak at 405 nm. In a set period of time, the higher the absorbance of the mixture of pNPB and enzyme solution is at 405nm, the higher the yield of p-nitrophenol and the greater the enzyme activity.
Based on the results , we found that both RIDD-PETase and RIDD-MHETase have esterase activity. After 20 minutes of reaction, each resulted in absorption peaks as high as 3.4873 and 2.7654 at 405nm.
PET plastic sheet degradation test
We obtained PET plastic sheet with 12% crystallinity ( scientific research only) from TJUSLS_China, and cut it into 5mm*5mm fragments (0.07g per fragment). We incubated 1.5 mL of E.coli BL21, E.coli BL21/pET28a-PD, E.coli BL21/pET28a-M, E.coli BL21/pET28a-PD-MD-hA cell pellet and medium in several EP tubes,and added three 5mm*5mm fragments, respectively. We then incubated the reaction mixture at 37℃ for 7 days. After 7 days, the degradation product, erephthalic acid (TPA), was detected by UV Spectrophotometry and thus determined PET degradation efficiency.
In terms of measurement, we chose to use UV spectrophotometry to detect the output of TPA. Binding with RIDD-PETase and RIDD-MHETase, PET will be decomposed with ethylene glycol (EG) and terephthalic acid (TPA) as final product. Ethylene glycol is volatile, and the test results are not credible, so we decided to detect TPA. Through previous literature research, we found that there are two mainstream detection methods for TPA. One is to directly perform UV spectrophotometry on the sample. The increase in absorbance of the reaction mixture in the ultraviolet region of the light spectrum (at 240 nm) indicates the release of soluble TPA or its esters from an insoluble PET substrate. This compound shares an identical strong absorbance peak around 240–244 nm with an identical extinction coefficient as all three compounds contain the same number of carbonyl groups. The second is to adopt reverse-phase HPLC. Reverse-phase HPLC systems have been widely used to analyze the products derived from the enzymatic hydrolysis of PET owing to their powerful resolving capability and reproducibility. The different compounds produced by PET hydrolytic enzymes (i.e., TPA, MHET, and BHET) can be efficiently separated on a C18 reverse-phase HPLC column: The reaction mixture is loaded into a column equilibrated with a polar mobile phase and the concentration of the organic solvent (acetonitrile).
Considering our experimental cycle, throughput, and laboratory conditions, we chose UV spectrophotometry to detect TPA. Then we made the standard curve of TPA at OD240 (Figure 3) .
The liquid obtained after incubation of the above eight samples was tested for TPA content, and the blank absorption was subtracted. The data obtained is shown in Figure 4.
It can be seen from the concentration of TPA product that the concentration of TPA in PD-MD-hA’s medium is higher than that in PD’s medium or MD’s medium. This strongly supported the engineering success of our three-enzyme complex construction.
Scanning electron microscopy
To further confirm the activity of the three-enzyme complex we constructed, we also selected PET plastic sheets in E.coli BL21, E.coli BL21/pET28a-PD, E.coli BL21/pET28a-M, E.coli BL21/pET28a-PD-MD-hA medium to be examined by scanning electron microscopy The results were consistent with expectations. The plastic sheet treated with either E.coli BL21 medium or MD medium has almost no scratches or holes. The plastic sheet after PD treatment has some obvious scratches, while the plastic sheet after PD-MD-hA treatment showed surface covered with scratches, and at the same time, densely packed with holes of various shapes. This scanning electron microscopy further proved that the three-enzyme complex we constructed has a better PET plastic degradation activity than single-enzyme degradation.
Briefly, we successfully constructed three-enzyme complex as expected, measured the complex's enzyme activity, and performed scanning electron microscopy and TPA detection experiments. The degradation effect of the three-enzyme complex on PET plastic was evaluated qualitatively and quantitatively.
Through the qualitative test of scanning electron microscopy, the enzyme complex created in our project has better effect on PET plastic sheets than PETase alone. The use of PETase alone will only cause scratches on the surface of the plastic sheet. However, our enzyme complex has exceeded PETase activity, causing more significant scratches and on top of that, producing many deep holes. The results of scanning electron microscopy also indirectly accounted for the role of hydrophobin 4 in the enzyme complex. With the presence of hydrophobin 4, the enzyme complex can bind to the surface of the PET plastic sheet. Therefore, instead of dispersing in water and induce reaction through random collisions, the enzyme is more likely to dig deeper into the surface.
Through the quantitative test of TPA production detection, we found that the degradation effect of our enzyme complex is better than PETase used alone, and the degradation efficiency has been increased by two times.
Overall, we successfully constructed a three-enzyme complex, and confirmed its activity by analysis of degradation effect and product yield . It will be promising for the subsequent enzyme complex to efficiently degrade PET plastic and the industrialization of PET plastic bio recycling.
| None |