Part:BBa_K3859007
crRNA for GBSZ ARTAG barcode
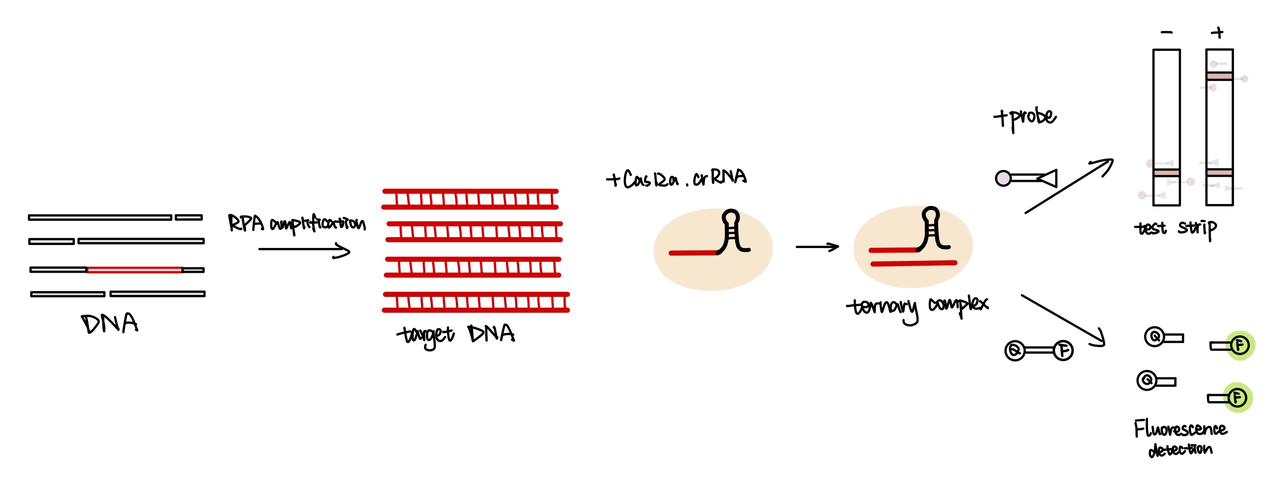
BBa_K3859007 is crRNA we designed for CRISPR Cas12a detection. crRNA is a guied RNA for cas12a enzyme to find the target sequence and combine to it. It made of two components one is repeat sequence and another one is target sequence[1]. The crRNA repeat, which is "UAAUUUCUACUAAGUGUAGAU" and will be the same for all EnGen Lba Cas12a guides. The repeat is followed by a target specific sequence, which will vary depending on the target. The target sequence of this crRNA(BBa_K3859007) is barcode GBSZ ARTAG(BBa_K3859000). We transfer the BBa_K3859000 which is a segment of DNA sequence into RNA sequence and add to the repeat sequence(fig.1).
Characterization
The crRNA is designed to be complementary to the target DNA sequence. So when the cas12a-crRNA complex is added to the system, the crRNA will recognize the target DNA sequence and bind with it, forming a ternary complex. Then the cas12a protein will start to cut the single strand DNAs (ssDNA)-it will cut not only on the target sequence, but also any single strand DNA that presents in the system. As a result, the ssDNA probe added into the reaction system will be cut by the cas12a protein. Depending the type of ssDNA probe added into the system, the probe will either produce florescence or show a red line on the test stripe(fig.2)[2].
Experienment and result
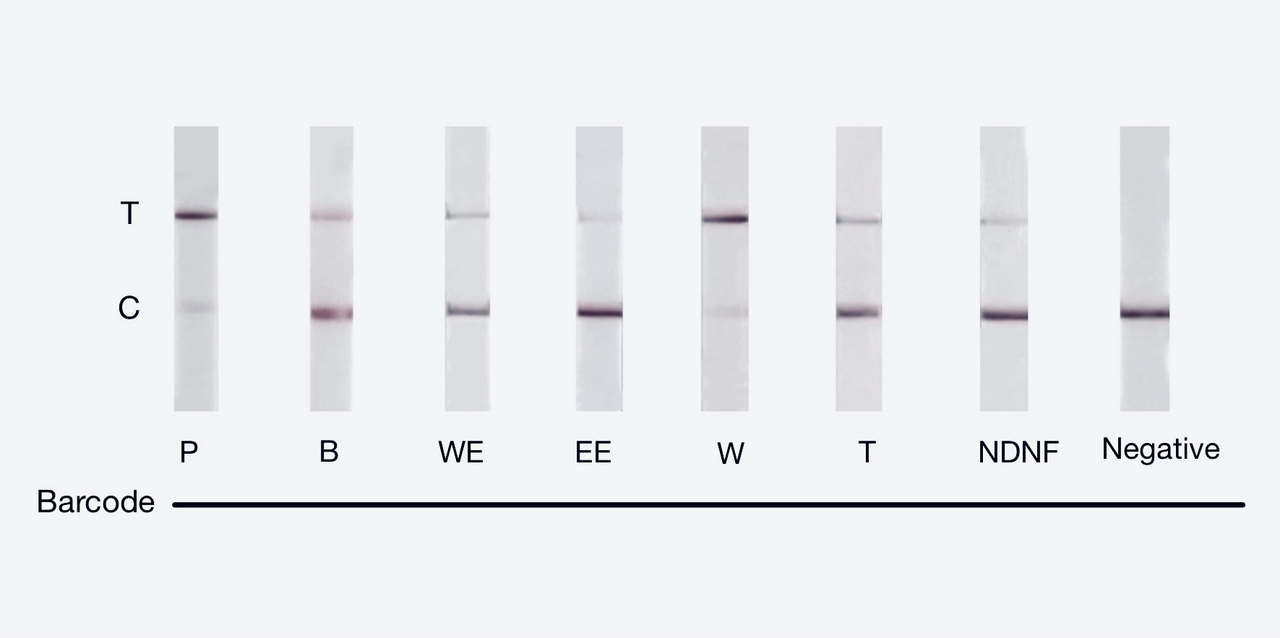
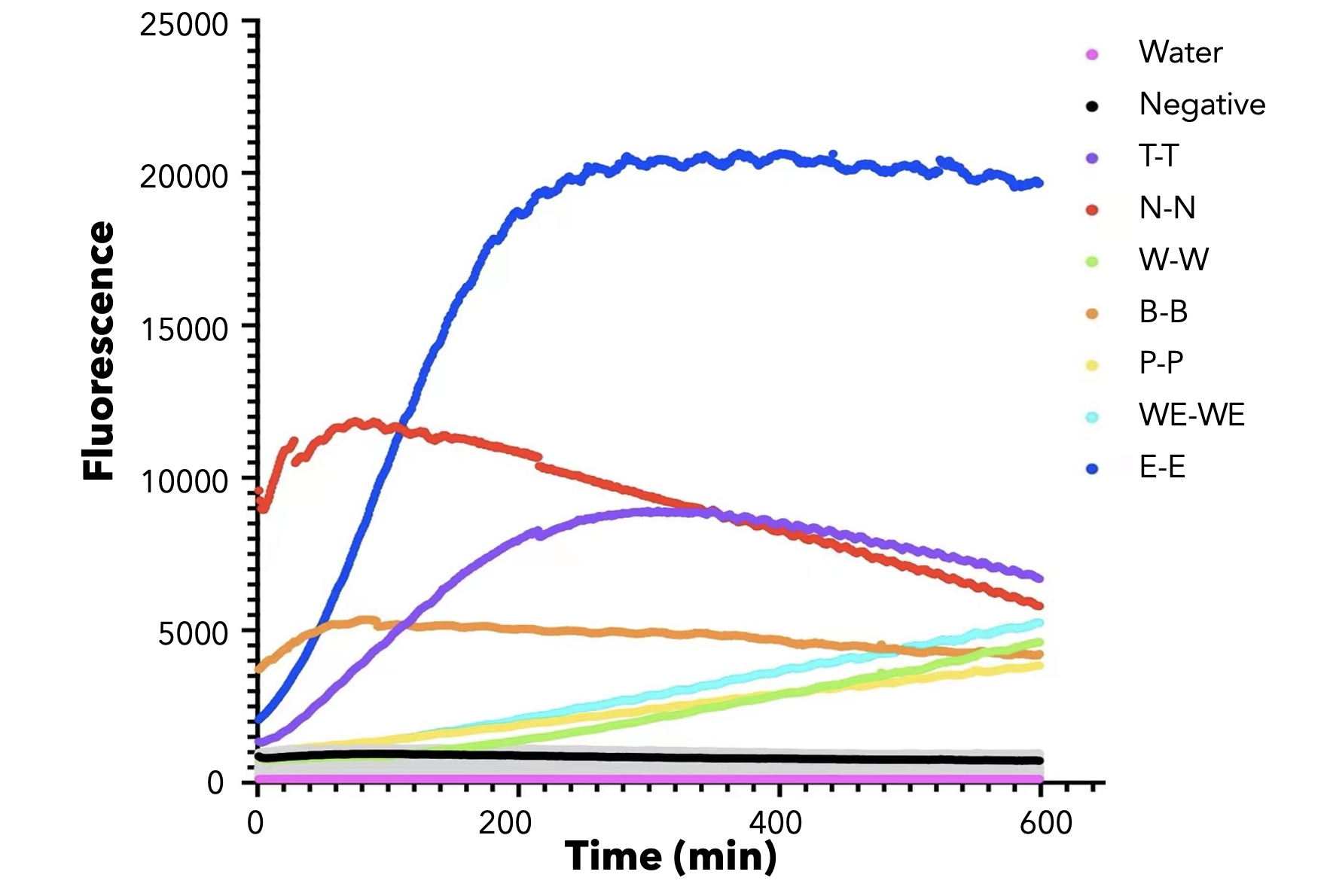
We used both the test strip and the florencence method. For the test strip, we successfully got the correct result for all the barcodes(fig.3). For the florencence method, we got the graphs of the florencence against time(fig.4)[2], which were proofed right by our model.
【fig.3】Cas12a test strip result
【fig.4】Cas12a fluorescence detection result. On the left of the symbol "-" refers to barcode name; On the right of the symbol "-" refers to crRNA name. All results for barcode mismatch with crRNA are shown in grey. E.g. "T-T" means the mixture of T-barcode and T-crRNA.
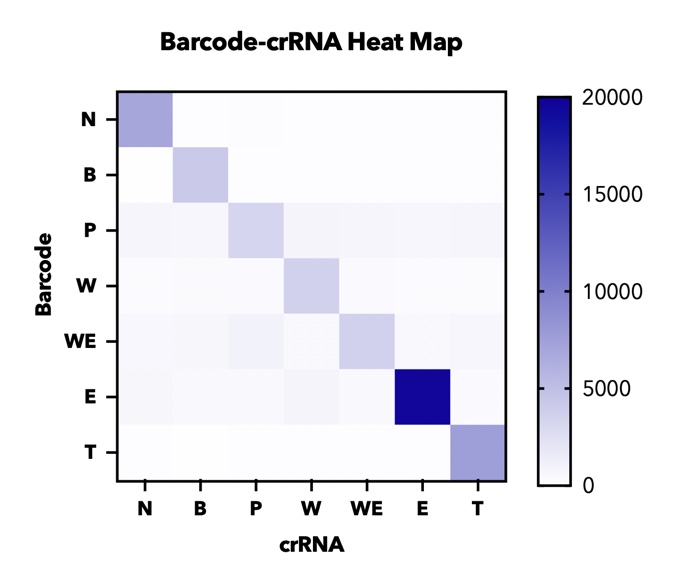
In addition, in order to test the specificity of our barcode design. We constructed 7 barcodes and their matching CRISPR RNAs (crRNAs) and assayed all permutations in vitro using cas12a florencence detection(fig.5)[2].
【fig.5】Heatmap of endpoint fluorescence values from in cas12a fluorescence detection of all combinations of 7 barcodes and 7 crRNAs assessing specificity of each barcode-crRNA pair.
References
1. Li, S. Y., Cheng, Q. X., Liu, J. K., Nie, X. Q., Zhao, G. P., & Wang, J. (2018). CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell research, 28(4), 491–493. https://doi.org/10.1038/s41422-018-0022-x
2. Qian, J., Lu, Z. X., Mancuso, C. P., Jhuang, H. Y., Del Carmen Barajas-Ornelas, R., Boswell, S. A., Ramírez-Guadiana, F. H., Jones, V., Sonti, A., Sedlack, K., Artzi, L., Jung, G., Arammash, M., Pettit, M. E., Melfi, M., Lyon, L., Owen, S. V., Baym, M., Khalil, A. S., Silver, P. A., … Springer, M. (2020). Barcoded microbial system for high-resolution object provenance. Science (New York, N.Y.), 368(6495), 1135–1140. https://doi.org/10.1126/science.aba5584
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |