Part:BBa_K3743014
Cas12g
Part Description
RNA-guided Cas12g is a ribonuclease that mainly targets single-stranded RNA substrates. CRISPS-Cas12g recognizes RNA substrates specifically, Comparing it to other Cas12 proteins that have been identified so far, which makes it a potentially useful platform for transcriptome engineering and diagnostics.
A bilobed structure of Cas12g shows a miniature NUC2 and REC2 domain, whereas the guided RNAs fold into an "F" shaped structure recognized mainly by the Rec lobes. Target RNA and crRNA guide form a duplex to insert into the central cavity between the REC and NUC lobes, altering their conformation and activating Cas12g. this insights would make the development of Cas12g applications much easier.
Usage
The Gly Ser linker is utilised to conjugate it with the L7Ae protein (which inhibits transcription by binding to its kink-turn). The mechanism simply identifies and attaches to mRNA in the cancerous environment, causing the L7Ae protein to be consumed. As a result of not binding to kink-turn, the inhibitory action on transcription is inhibited, resulting in transcription activation and a rise in vaccine yield. As a result, it is a cell-specific design that binds to PD-L1 mRNA, which plays an immune evasion role in cancerous environments, particularly TLCs.
Improvement by AFCM-Egypt 2022 team
This part improved by AFCM-Egypt 2022 team in this part BBa_K4140016
Improvement by adding kink-turns
This year we employed this approach in our circuit as a regulatory domain, as we took advantage of Cas12g properties to control the expression of PAH in our therapeutic circuit as it differs from other Cas12 proteins by targeting a single-strand RNA substrate making it a potent platform for post-transcriptional modification, and reduces the chance of gene knockdown as it inhibits translation not the transcription by nucleotide deletion like other Cas protein due to inactivation of its NUC domain which mediates the nuclease activity of Cas protein. We carried out an improvement to its functionality limiting its off-targeting effect by adding a kink turn upstream to the 5’ end of its mRNA (translation initiation site) as shown in figure 1, which acts as a translation switch for Cas12g by repressing its synthesis in the presence of L7Ae due to the formation of a crystal structure composed of L7Ae-boxC/D k-turn complex which inhibits ribosomal function on the mRNA of Cas12g but in the absence of L7Ae protein Cas12g is freely expressed. The presence or lack of L7Ae is correlated with the concentration of phenylalanine since TyrR dimer, ATP, and the paroF promoter are required for L7Ae's production.
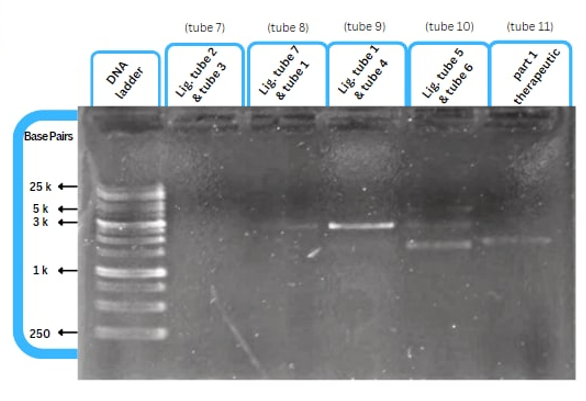
Experimental validation of the improvement by AFCM-Egypt 2022 team
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 3. The running part (ordered from IDT) included KP-SP and lacZ alpha.

This figure on the left (Phe-ve/X-gal +ve) shows that the WCB emits a low signal of the blue color at 0 mg/dL of phenylalanine. The results are shown to the right (Phe-ve/X-gal +ve), which shows that the WCB emits a negligible signal of the blue color at 0 mg/dL of phenylalanine. which is also a background noise from the circuit but with a very low signal after addition of the regulatory circuit. This iteration was essential as it proved the concept of background noise without using the regulatory circuit unless the result after transforming it.

For the previous part without improvement, the plate showed an incomplete negative percentage of dominant control color in the test sample. To confirm the results, we plotted the absorbance (nM) of both the tested sample and the control sample (standard positive) as they showed slight overlapping ranges of wavelength (nM).
For the improved part after transforming the regulatory circuit (kink turn- cas12g), the plate showed a negative percentage of dominant control color in the test sample. To confirm the results, we plotted the absorbance (nM) of both the improved sample and the control sample as they showed different ranges of wavelength (nM).
Improvement by directed evolution
After performing mutagenesis prediction of mutational landscape of cas12g and tested the effect of these mutations on the evolutionary fitness of the protein after generating multiple sequence alignment of the protein sequence and predict mutational landscapes. As shown in the chart, the (R627W) mutation showed the highest score compared to other mutations. On the contrary, we can see that the (L654F) contributed to the lowest evolutionary fitness to cas12g.As shown in Figure (5)
Literature Characterization
A Cas12g–sgRNA–target RNA ternary complex was assembled by incubating a catalytically inactive Cas12g (E655A) with the sgRNA and a 24-nucleotide target RNA in order to obtain insight into target RNA recognition. By using cryo-EM, we were able to determine the structure with a 4.8 resolution. As shown in figure (1). The nuclease activity was abolished when REC1220–354 was deleted, showing that it plays a crucial role in substrate cleavage (Figure 2). The positively charged residues in the REC1220–354 region are obviously involved in the recognition of the crRNA–target RNA duplex. Mutation of six positively charged residues within the first 20 amino acids of this region (K221, K223, R224, R226, R232, and K237) had a comparable effect on RNA cleavage efficiency as deletion of REC1220–354 in support of this statement.(1)
References
1.Li, Z., Zhang, H., Xiao, R., Han, R., & Chang, L. (2021). Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nature Chemical Biology, 17(4), 387–393.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 313
Illegal PstI site found at 787
Illegal PstI site found at 1729 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 313
Illegal PstI site found at 787
Illegal PstI site found at 1729 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1308
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 313
Illegal PstI site found at 787
Illegal PstI site found at 1729 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 313
Illegal PstI site found at 787
Illegal PstI site found at 1729
Illegal NgoMIV site found at 1230 - 1000COMPATIBLE WITH RFC[1000]
| None |