Part:BBa_K3743004
MS2 (DD) by fusion
Part Description
The destabilization domains (DD) show a fusion protein component that is unstable and destabilizes other proteins, leading to protein degradation. A well-known example of DD is mutated DHFR (E. coli) (DDDHFR), which causes the degradation of proteins in cells. However, if a trimethoprim analog (TMP) is added to bind the destabilizing domain, the fusion protein can stabilize and return the expression to normal levels. This approach allows the regulation of secreted proteins and their biological activity, allowing functional control over proteins of interest in a variety of contexts in mammalian cell cultures, the Plasmodium and Toxoplasma apicomplexans, and in live mice, creating new avenues to open various applications such as cancer therapy and targeted gene delivery. In our part, the destabilizing domain binds to the ms2 coat protein that inhibits it from binding to its riboswitch until trimethoprim analog (TMP) is added which acts as a small molecule inhibitor will free the MS2 after stabilizing DD. Free MS2 inhibits the circuit to be able to control the transcription process in extreme conditions as cytokine storm or unpredictable results.
Usage
This part represents the final form of the used riboswitch to be controlled by Trimethoprim (TMP) that has a small molecule inhibitor role. Destabilizing domain (DD) is fused to MS2 by Gly-Ser linker. And this is an improvement of both parts. If the vaccine is uncontrollable, TMP will be administered that will stabilize DD therefore, binding of MS2 to U2-snRNP that finally inhibits the circuit and stops the transcription in extreme conditions.
Literature Characterization
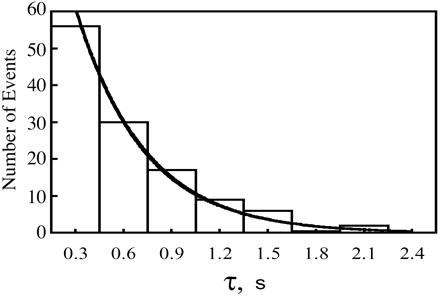
In one study that has done an exponential fit of the data as shown in figure (1). The study made a summary the ensemble averaged rate constants at various methotrexate concentrations with a limited degree of uncertainty due to the limited number of events and limited time resolution of the equipment.(1)
Characterization Of Mutational Landscape
A mutational landscape prediction through saturation mutagenesis of MS2-DD protein and the effect of these mutations on the evolutionary fitness of the protein is tested after generating multiple sequence alignment of the protein sequence and predict mutational landscapes. As shown in the chart, the (W70T) mutation showed the highest score compared to other mutations. On the contrary, we can see that the (L74G) contributed to the lowest evolutionary fitness to MS2-DD.As shown in Figure (2)
Characterization by Mathematical Modelling
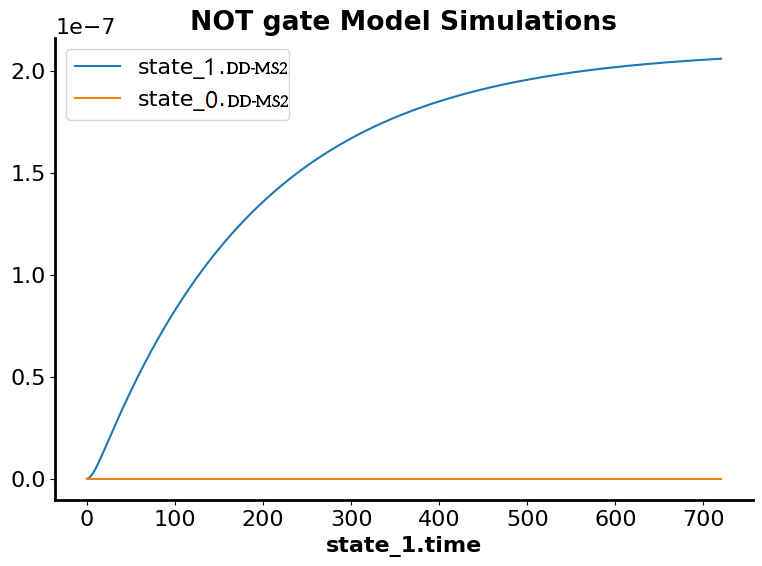
NOT Gate
In the simulation of mass action kinetics of DD_MS2 which is controlled by TMP, the blue plot represents that the steady state is achieved after 700 seconds in the absence of TMP and the red plot represent the inhibition of the transcription through binding of MS2 to small nuclear ribonucleoprotein (snRNP) which is controlled by TMP to terminate the transcription if the circuit is uncontrollable.showed increased transcription of the circuit which reach the steady state after about 200 seconds.
AND Gate
Blue plot achieves the maximum concentration at 1.6 after 700 seconds which represents the presence of 2 inputs in the AND (absence of TMP and binding of miRNA to toeholdOn switch upstream to the vaccine) and the red plot represents the inhibition of the transcription in absence of one of the two inputs wither the presence of the TMP so stabilized the destabilizing domain and inhibiting the circuit or absence of binding of miRNA to ToeholdOn switch
References
1.Rajagopalan, P. R., Zhang, Z., McCourt, L., Dwyer, M., Benkovic, S. J., & Hammes, G. G. (2002). Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics. Proceedings of the National Academy of Sciences, 99(21), 13481-13486.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 292
| None |