Part:BBa_K3478889
pTAC-GPPS-bLIS
.
Description
GPPS-bLIS is composed of two coding sequences, GPPS (https://parts.igem.org/Part:BBa_K2753002) and bLIS (https://parts.igem.org/Part:BBa_K3478890). Coding GPPS is used to produce Geranyl pyrophosphate synthase that produce GPP from DMAPP, which is a substrate for linalool synthase that produces linalool. in our experiment, we constructed pR6K-ptac-GPPS-bLIS to produce linalool. The vector of the part is pR6K, with the promoter ptac, and composite part GPPS-bLIS. We first amplified the gene fragments using PCR (figure 1A, B, C and D) , and purified them to get pure gene fragments for pR6K-ptac-GPPS and bLIS.
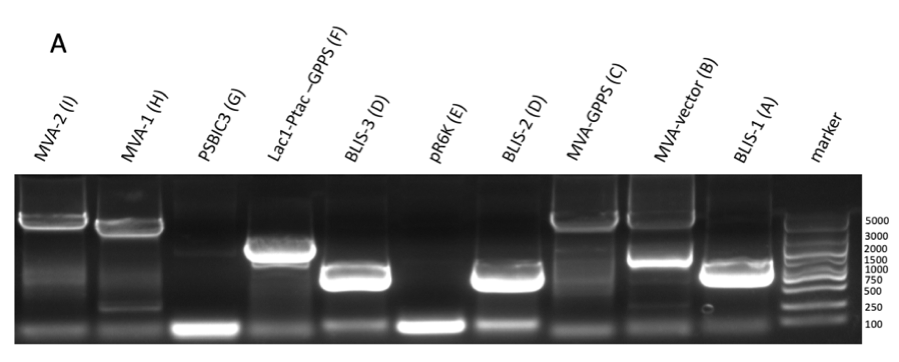
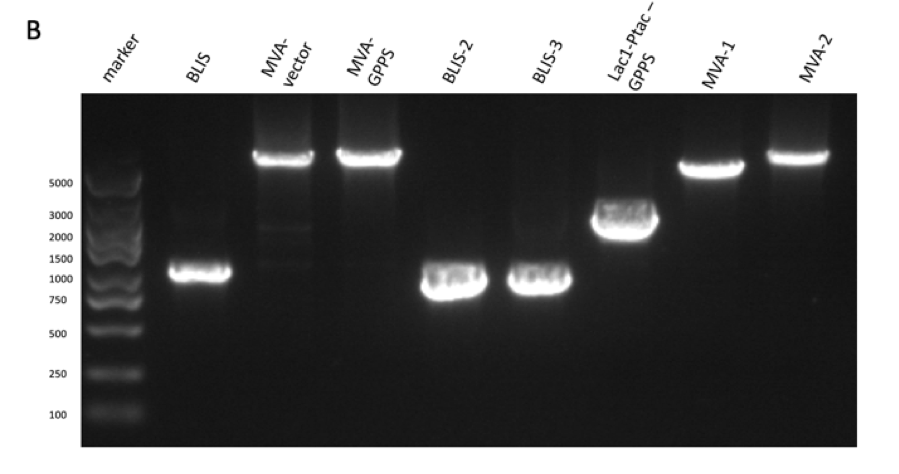
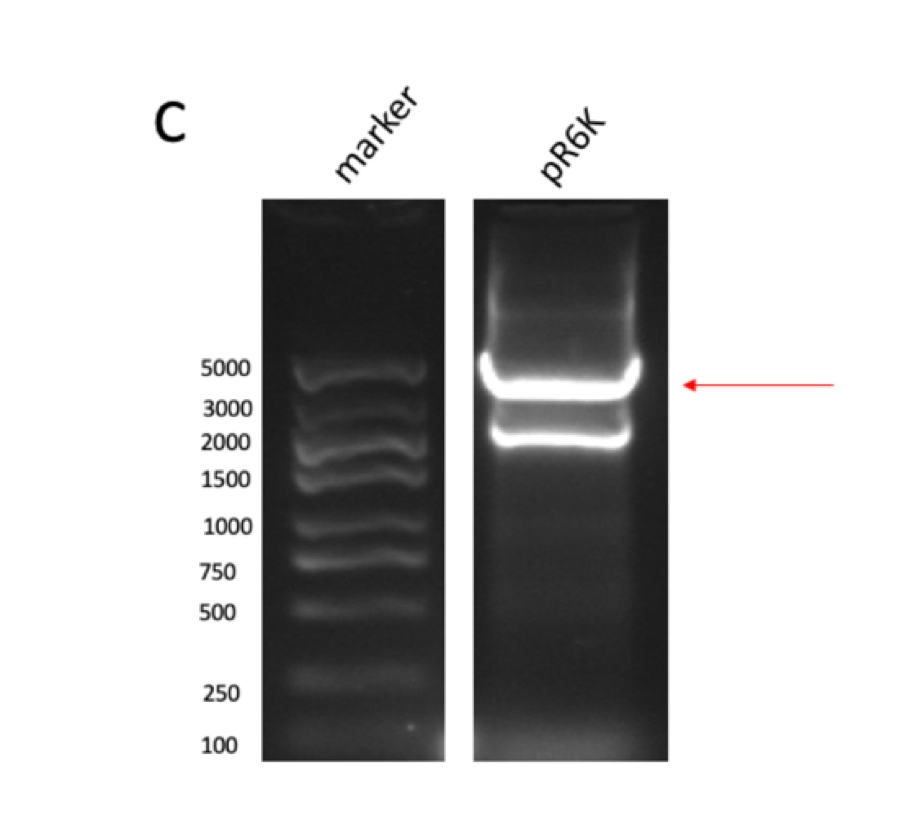
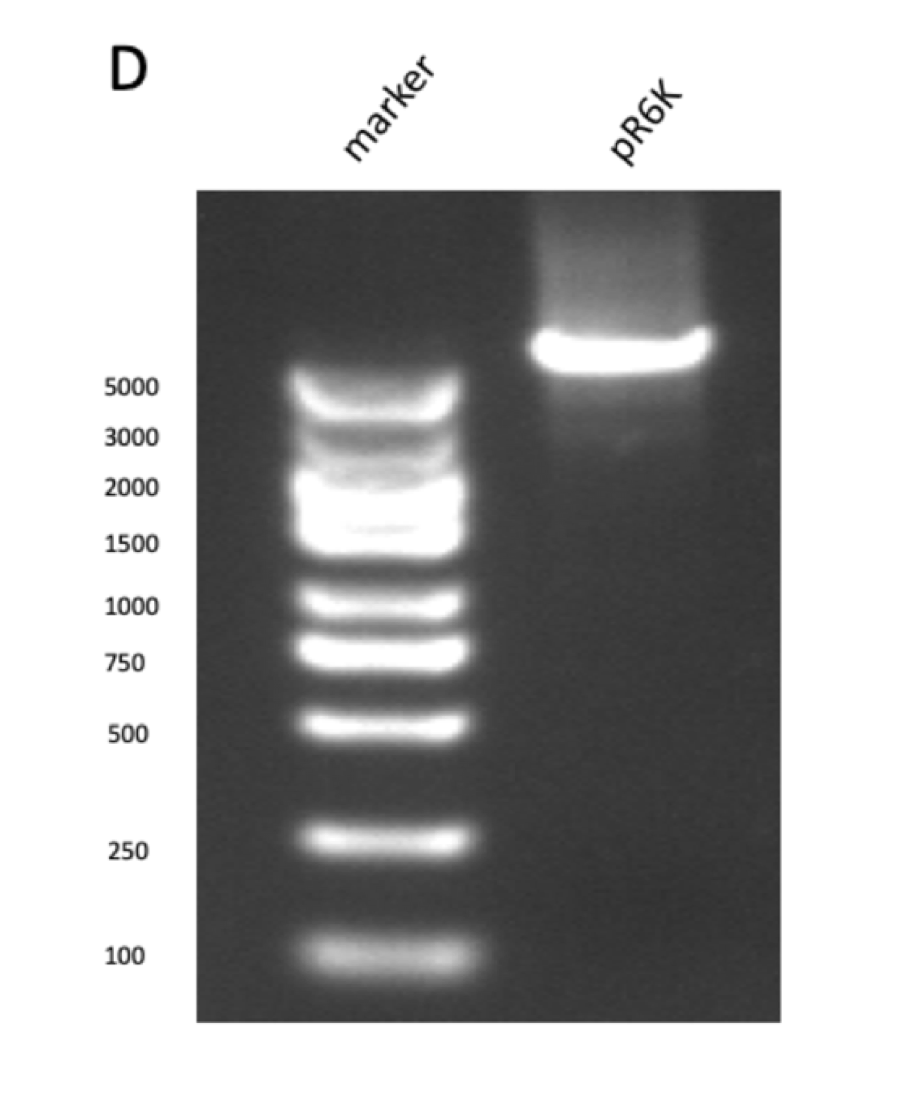
Figure figure 1: the gel DNA gel electrophoresis results. (A) amplification result of gene segments of p15A-MVA-ptac-GPPS-bLIS (BLIS-1 (1015 bp), MVA-vector (6372 bp), MVA-GPPS (6970 bp)), pR6K-ptac-GPPS-bLIS (BLIS-2 (1016 bp), pR6K (5958 bp)) , pSB1K3-ptac-GPPS-bLIS (BLIS-3 (1016 bp), Lac1-Ptac-GPPS (2371 bp), PSBIC3 (2111 bp)) , p15A-MVA (MVA-1 (5177 bp), MVA-2 (6922 bp)). (B) result of gene segment purification. BLIS-1 (1015 bp), MVA-vector (6372 bp), MVA-GPPS (6970 bp), BLIS-2 (1016 bp), BLIS-3 (1016 bp), Lac1-Ptac-GPPS (2371 bp), (MVA-1 (5177 bp), MVA-2 (6922 bp). (C) amplification result of pR6K (5958 bp). (D) purification result of pR6K (5958 bp).
Gibson assembly was used to create plasmid pR6K-patc-GPPS-bLIS. We transformed the pR6K-patc-GPPS-bLIS to E. Coli DH5 α. The success of transformation is verified by a colony PCR. And induced it with IPTG, which allows it to produce linalool synthase (figure.2)
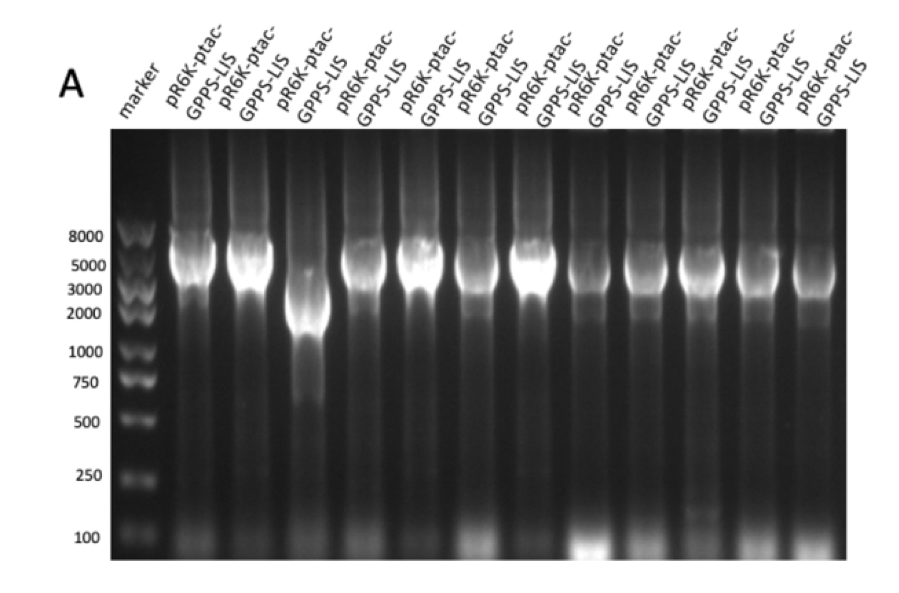
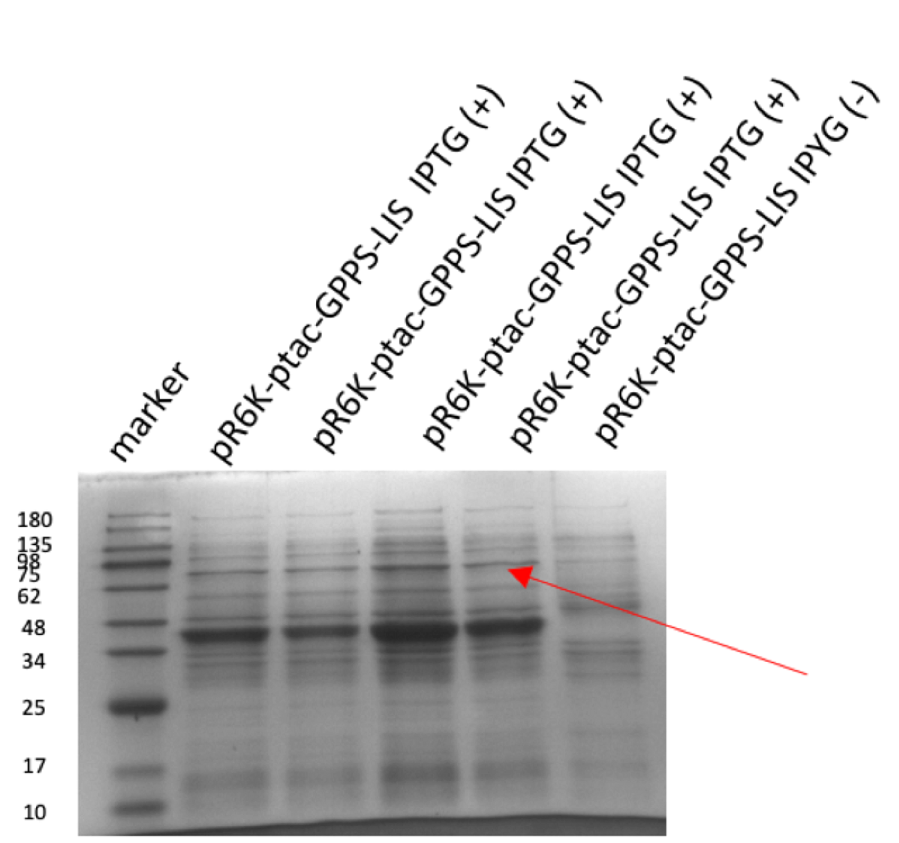
Figure 2: (A) gel electrophoresis result of pR6K-ptac-GPPS-bLIS (14358 bp). (B) protein electrophoresis results of linalool synthase (65.6 kDa) However, pR6K-patc-GPPS-bLIS alone in the E. Coli can’t produce linalool, because the substrate of GPP synthase, DMPP, is produced by MVA pathway, so we also constructed a plasmid p15A-MVA (figure 3), which allows the bacteria to have MVA pathway in it and produce DMPP, so that linalool synthesize can be achieved.
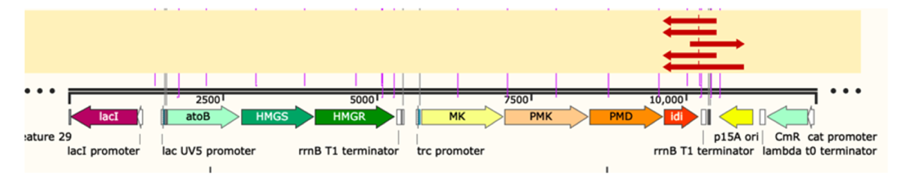
Figure 3: gene sequencing result of p15A-MVA.
We transformed the two plasmid in We cotransformed P15A-MVA and pR6K-ptac-GPPS-bLIS plasmid to E.coli DH5α. we induced them with IPTG and two of them with extra glucose. after the inducing process, the bacteria solution was treated with n-hexane to extract purified linalool(figure 4). We can clearly smell the strong fragrance of linalool in those samples, and samples with glucose have a stronger fragrance than samples that are not treated with glucose. This indicates that glucose may promote the synthesis of linalool in E.coli DH5α. We do not need to knock out the gene in E.coli producing stinky smell because the aroma overcomes the smell of E.coli. Therefore, our E.coli won’t affect the local environment and appearance.
Besides, we also compared the smell of our sample to standard linalool sample, and standard geraniol, which is the product of pR6K-ptac-GPPS-GES (the plasmid before our improvement) Based on our observation the aroma of geraniol contains sweetness; whereas, the aroma of standard linalool sample has a slight peppery smell. The two smells are quite distinguishable. The small of our samples also have a slightly peppery aroma, which is close to the linalool standard sample. Thus, we have successfully produced linalool. In addition, through visual observation, we also confirmed that glucose can promote the synthesis of linalool in E.coli. The transparent liquids in the test tubes are purified linalool (figure 4). The volume of the liquid in samples treated with glucose is larger than samples that aren’t treated by glucose. Therefore, E.coli treated with glucose may be able to produce more linalool in the same condition thanE.coli that is not. Our next step may be finding the optimal condition for linalool synthesis in E.coli. However, due to the Covid-19 situation, we do not have enough time in the lab, so we can only do those experiments in the future.
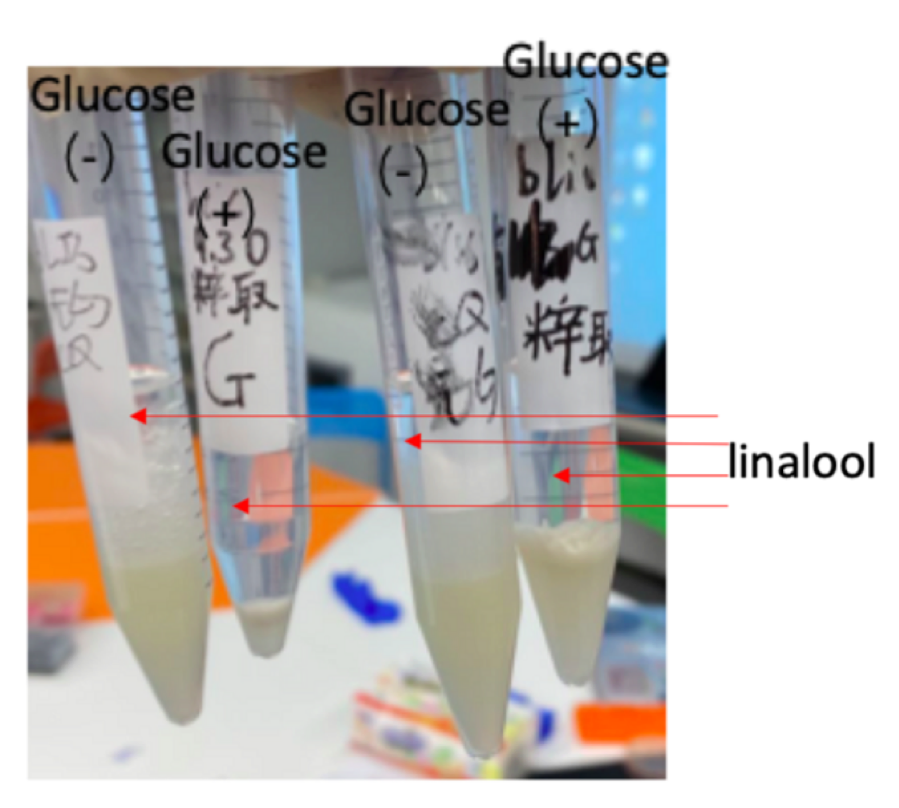
Figure 4: E.coli DH5α with P15A-MVA and pR6K-ptac-GPPS-bLIS was propagated in four separated conical flasks. After the bacteria solutions reach the suitable OD (0.6-0.8), we added 2.5 ml glucose to two of the mediums during and added 25mM of IPTG to By adding n-hexane (4.5ml) to the induced bacteria solution, we can extract pure linalool (transparent layer)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2955
Illegal AgeI site found at 2534
Illegal AgeI site found at 3210 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 2893
| None |
