Part:BBa_K3447103
Validation of Arabinose response system
This part contains an arabinose operon, which constitutes the arabinose sensing system. By connecting the sfGFP gene downstream, we were able to verify the expression intensity of this arabinose operon.
Usage and Biology
Arabinose operon is composed of AraC protein, PBAD and PC. AraB, araA and araD are three genes required for arabinose degradation in E. coli. They form a gene cluster, abbreviated as araBAD, which encodes three enzymes required for arabinose metabolism: Ribulose kinase, Arabinose isomerase and ribulos-5-phosphate eimerase.
The expression of araBAD gene is regulated by araC transcription factor, the fourth gene in the Ara operon. AraBAD and araC genes are transcribed in opposite directions on the two strands. AraBAD is translated from PBAD to the right and araC from PC to the left.
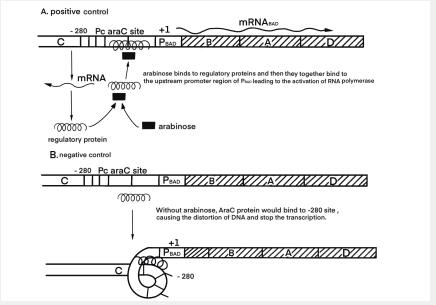
As shown in Figure 1, when arabinose is present in cells, araC, the product encoded by gene araC, binds to arabinose and binds to the upstream promoter region of PBAD to activate RNA polymerase and activate the gene after PBAD. On the contrary, when arabinose is not present, the product araC will cause DNA curling to inhibit the binding of RNA polymerase, and inhibit the expression of gene downstream of PBAD.
Characterization
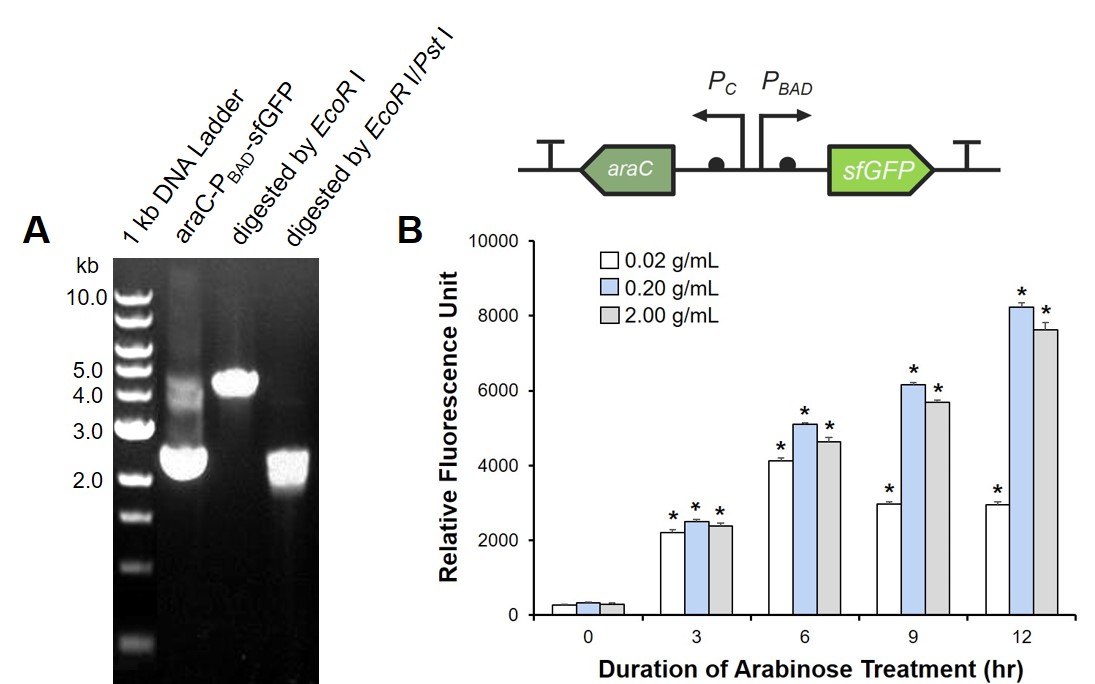
To prove the arabinose sensing system can work successfully, a sfGFP was inserted after PBAD under the induction of arabinose at a series of concentrations. As shown in Fig. 2B, the expression efficiency was highest when induced with 0.2 g/mL arabinose.
Design
Design Notes
We added some synonymous mutations to avoid part rules.
Source
We found this sequence data in the previous iGEM teams (Glasgow 2017) and in GenBank.
References
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1216
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1155
Illegal XhoI site found at 1666 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 990
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1
Illegal SapI site found at 972
| None |